generic_driver.h File Reference
Header file for Generic Driver. More...
#include "globals.h"
Go to the source code of this file.
Defines | |
| #define | numRunSets 150 |
Functions | |
| void | get_parmf () |
| Get user's run-time parameters from configuration file. | |
| void | setup () |
| Controller to call functions to set up & initialize the model. | |
| void | track_time (int istep) |
| Keep track of simulation time. | |
| int | PTSL_test () |
| Test for presence of point-time series meteorological data. | |
| int | ModExperim_test () |
| Test for presence of special parameter file for model experiments. | |
| ViewParm * | read_output_parms (void) |
| Call the read_output_parms function. | |
| void | read_model_parameters (char *s_parm_name, int s_parm_relval) |
| Call functions to read model parameters from datafiles. | |
| void | getInt (FILE *inFile, const char *lString, int *iValPtr) |
| Get an integer following a specific string. | |
| void | getFloat (FILE *inFile, const char *lString, float *fValPtr) |
| Get a float value following a specific string. | |
| void | getString (FILE *inFile, const char *lString, char *inString) |
| Get a string following a specific string. | |
| void | calcdate (double jd, int *m, int *d, int *y, int *h, int *mi, double *sec) |
| Determine the Gregorian date from a Julian calendar day. | |
| void | setup_platform () |
| Effectively unused in serial (non-parallel) implementation. | |
| void | set_env_vars (void) |
| Acquire necessary environment variables. | |
| void | setup_grid () |
| Provide the 2D array size for the model domain. | |
| int | skip_white (FILE *infile) |
| Skip white space(s) in a file. | |
| int | scan_forward (FILE *infile, const char *tstring) |
| Scan forward until a particular string is found. | |
| double | julday (int mon, int day, int year, int h, int mi, double se) |
| Determine the Julian calendar day from a Gregorian date. | |
| float | FMOD (float x, float y) |
| Modulus of a pair of (double) arguments. | |
| void | local_setup (int argc, char **argv) |
| Does very little in serial implementation (opens a low-level debug file). | |
| void | open_debug_outFile (int index) |
| Open debug-related output files. | |
| void | broadcastInt (int *iValPtr) |
| Parallel code: does nothing in serial implementation). | |
| void | sync_processors () |
| Parallel code: does nothing in serial implementation). | |
| void | send_point_lists2 (SeriesParm *pSeries, int nSeries) |
| Point time series output: print time series data to file(s). | |
| void | open_point_lists (SeriesParm *pSeries, int nSeries) |
| Point time series output: open files and print headers. | |
| void | init_static_data (void) |
| Initialize static spatial data. | |
| void | init_dynam_data (void) |
| Initialize dynamic spatial data. | |
| void | init_eqns (void) |
| Initialization of the model equations. | |
| void | init_canals (int runNumb) |
| Call to initialize the water managment canal network topology and data. | |
| void | init_succession (void) |
| Call to initialize the habitat succession module. | |
| void | PtInterp_read (void) |
| On-the-fly interpolations to develop spatial time series from point data. . | |
| void | reinitBIR (void) |
| Calls to re-initialize Basin/Indicator-Region data. | |
| void | reinitCanals (void) |
| Call to re-initialize canal storages. | |
| void | alloc_memory () |
| Allocate memory. | |
| void | gen_output (int step, ViewParm *view) |
| Generate output. | |
| void | get_map_dims (void) |
| Get the map dimensions of the global model array. | |
| int | call_cell_dyn (int sector, int step) |
| Calling function for the cell_dyn** dynamic ecological modules. | |
Variables | |
| int | NSector |
| int | iSector [MAX_SECTOR] |
| int | seed |
| int | ESPmodeON = 0 |
| int | WatMgmtOn |
| int | HabSwitchOn |
| int | PositAnalOn = 0 |
| int | IsSubstituteModel = 0 |
| char | SimAlt [30] |
| char | SimModif [30] |
| char | outpath [120] |
| int | gbl_size [2] |
| char | initDateRead [15] |
| double | Jdate_init |
| double | Jdate_end |
| int | yr_in |
| int | mo_in |
| int | da_in |
| int | hr_in |
| int | mi_in |
| int | se_in |
| int | yr_end |
| int | mo_end |
| int | da_end |
| int | mo_R_in |
| int | da_R_in |
| int | PORnumday |
| int | N_iter |
| int | istep |
| float | step_Cell |
| float | avg_Intvl = 0 |
| float | budg_Intvl = 0 |
| float | BIRavg_Intvl = 0 |
| float | BIRhyd_avg_Intvl = 0 |
| float | can_Intvl = 0 |
| int | budgCalendar |
| int | BIRhydCalendar |
| int | avgCalendar = 0 |
| int | canalCalendar = 0 |
| int | avgPrint = 0 |
| int | canPrint = 0 |
| ProgAttr * | ProgExec |
| ProgAttr ** | RunList |
| float | gRTable [] |
| ViewParm * | view |
| SeriesParm | pSeries [MAX_PTSERIES] |
| Point2D | dbgPt |
| struct nodenv | env |
| int | procnum |
| int | Lprocnum |
| int | nprocs [2] |
| int | recpnum [2] |
| int | tramType |
| int | tramNum [2] |
| int | lcl_size [2] |
| int | lcl_start [2] |
| char * | ModelPath |
| char * | ProjName |
| char * | OS_TYPE |
| char * | OutputPath |
| char | modelName [20] |
| char | modelVers [10] |
Detailed Description
Header file for Generic Driver.
This defines or declares variables & functions that are global to Generic_Driver.c.
Note: documented with Doxygen, which expects specific syntax within special comments.
The Everglades Landscape Model (ELM).
last updated: Oct 2006
Definition in file generic_driver.h.
Define Documentation
| #define numRunSets 150 |
Definition at line 84 of file generic_driver.h.
Function Documentation
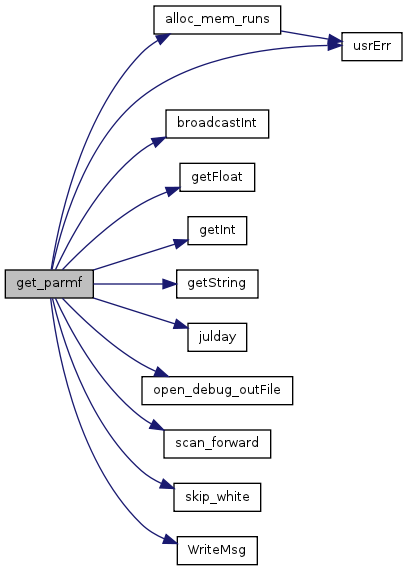
| void get_parmf | ( | ) |
Get user's run-time parameters from configuration file.
This is where the application is set up by the user's edits to the Driver.parm text configuration file, providing the simulation start and end dates, grid cell size, debug level, model scenario name, etc.
- Variables local to function
stop boolean flag to stop reading sector (celldyn module) numbers
ch character used in delineating the reading of sector (celldyn module) numbers/syntax
infile an input file pointer representing the Driver.parm file
filename the filename, including path, of the Driver.parm file
ss a string being parsed to read model configuration information from the Driver.parm file
Re_initDateRead data of simulation re-initialization (Position Analysis mode)
endDateRead date of end of simulation
Definition at line 288 of file Generic_Driver.c.
References alloc_mem_runs(), avg_Intvl, avgCalendar, BIRhyd_avg_Intvl, BIRhydCalendar, broadcastInt(), budg_Intvl, budgCalendar, can_Intvl, canalCalendar, canstep, CELL_SIZE, celWid, da_end, da_in, da_R_in, seriesParm::data, dbgPt, debug, dt, ESPmodeON, False, getFloat(), getInt(), getString(), gwstep, HabSwitchOn, hr_in, hyd_iter, initDateRead, iSector, Jdate_end, Jdate_init, julday(), MAX_PTSERIES, mi_in, mo_end, mo_in, mo_R_in, modelName, ModelPath, modelVers, msgStr, N_iter, NSector, open_debug_outFile(), outpath, OutputPath, PORnumday, PositAnalOn, procnum, ProgExec, ProjName, pSeries, RunList, prog_attr::S_ParmName, prog_attr::S_ParmRelVal, scan_forward(), se_in, sec_per_day, seed, SensiOn, sfstep, SimAlt, SimModif, skip_white(), sq_celWid, step_Cell, True, usrErr(), WatMgmtOn, WriteMsg(), point2D::x, point2D::y, yr_end, and yr_in.
Referenced by setup().
00289 { 00300 int i=0, stop=0; 00301 unsigned char ch; 00302 FILE *infile; 00303 char filename[120], ss[120]; 00304 char Re_initDateRead[15], endDateRead[15]; 00305 int SParmNum_TEMP = 150; /* TODO: will get this from reading the senitivity-parameter list */ 00306 00307 sprintf(filename,"%s/%s/RunParms/Driver.parm",ModelPath,ProjName); 00308 infile = fopen(filename,"r"); 00309 if(infile==NULL) { fprintf(stderr,"Error, can't open file %s",filename); exit(0); } 00310 00311 00312 fgets(ss,120,infile); 00313 sscanf(ss,"%s",outpath); 00314 OutputPath = (char*)&outpath; /* path for all output */ 00315 open_debug_outFile(0); /* opens the Driver0.out debug file */ 00316 00317 /* start date */ 00318 fgets(ss,120,infile); 00319 sscanf(ss,"%s",initDateRead); /* read in as yyyy/mm/dd format */ 00320 00321 sprintf(msgStr,"initDateRead=%s\n",initDateRead); WriteMsg(msgStr,1); 00322 sscanf(initDateRead, "%4d/%2d/%2d,",&yr_in,&mo_in,&da_in); /* starting date of sim */ 00323 00324 /* julian day, returns a double (args = month, day, year, hour, minute, second) */ 00325 Jdate_init = julday(mo_in, da_in, yr_in, hr_in, mi_in, se_in); 00326 00327 00328 /* end date */ 00329 fgets(ss,120,infile); 00330 sscanf(ss,"%s",endDateRead); /* read in as yyyy/mm/dd format */ 00331 00332 sprintf(msgStr,"endDateRead=%s\n",endDateRead); WriteMsg(msgStr,1); 00333 sscanf(endDateRead, "%4d/%2d/%2d,",&yr_end,&mo_end,&da_end); /* ending date of sim */ 00334 00335 /* julian day, returns a double (args = month, day, year, hour, minute, second) */ 00336 Jdate_end = julday(mo_end, da_end, yr_end, hr_in, mi_in, se_in); 00337 00338 /* re-initialization date */ 00339 fgets(ss,120,infile); 00340 sscanf(ss,"%s",Re_initDateRead); /* read in as mm/dd format */ 00341 00342 sprintf(msgStr,"Re_initDateRead=%s\n",Re_initDateRead); WriteMsg(msgStr,1); 00343 sscanf(Re_initDateRead, "%2d/%2d,",&mo_R_in,&da_R_in); /* re-initialize month/day */ 00344 00345 if (mo_R_in>0) { 00346 PositAnalOn=True; /* in position analysis mode, we re-initialize every year on same month/day */ 00347 sprintf(msgStr,"\n *** WARNING: Position Analysis capabilities are NOT VERIFIED FOR accuracy/consistency in ELMv2.3 - the Avg's, Basin/Indicator-Region output may not be reliable ***\n"); 00348 WriteMsg(msgStr,1); 00349 usrErr(msgStr); 00350 } /* TODO: fix the budget/stats re-inits for Position Analysis mode */ 00351 00352 fgets(ss,120,infile); 00353 sscanf(ss,"%s",modelName); 00354 sprintf(msgStr, "Model Name= %s", modelName); usrErr(msgStr); WriteMsg(msgStr,1); 00355 getString(infile,"Model version=",modelVers); /* model version number (eg., v.2.1) */ 00356 sprintf(msgStr,"Model version=%s\n",modelVers); WriteMsg(msgStr,1); 00357 00358 getFloat(infile,"CellArea=",&CELL_SIZE); 00359 sprintf(msgStr,"CellArea=%f\n",CELL_SIZE); WriteMsg(msgStr,1); 00360 celWid = sqrt(CELL_SIZE); /* cell width, used in number of hydro calcs */ 00361 sq_celWid = sqrt(celWid); /* square root of cell width, used in number of hydro calcs */ 00362 00363 00364 getFloat(infile,"budg_Intvl=",&budg_Intvl); 00365 sprintf(msgStr,"budg_Intvl=%f\n",budg_Intvl); WriteMsg(msgStr,1); 00366 budgCalendar = ( budg_Intvl == 0.0 ) ? (True) : (False); /* true/false flag signifying use of gregorian calendar for budget calcs 00367 a budg_Intvl of 0 days defaults to gregorian calendar-month 00368 intervals ( = #days/month, diff among months/years) 00369 In this case, the true/false budgCalendar flag is set to True */ 00370 00371 getFloat(infile,"BIRhyd_avg_Intvl=",&BIRhyd_avg_Intvl); 00372 sprintf(msgStr,"BIRhyd_avg_Intvl=%f\n",BIRhyd_avg_Intvl); WriteMsg(msgStr,1); 00373 BIRhydCalendar = ( BIRhyd_avg_Intvl == 0.0 ) ? (True) : (False); /* true/false flag signifying use of gregorian calendar for BIR hydro calcs 00374 a BIRhyd_avg_Intvl of 0 days defaults to gregorian calendar-month 00375 intervals ( = #days/month, diff among months/years) 00376 In this case, the true/false budgCalendar flag is set to True */ 00377 00378 getFloat(infile,"avg_Intvl=",&avg_Intvl); 00379 sprintf(msgStr,"avg_Intvl=%f\n",avg_Intvl); WriteMsg(msgStr,1); 00380 if ( avg_Intvl == 0 ) avgCalendar = True; /* true/false flag signifying use of gregorian calendar for recurring averaging calcs 00381 an avg_Intvl of 0 days defaults to gregorian calendar-month 00382 intervals ( = #days/month, diff among months/years) 00383 In this case, the true/false avgCalendar flag is set to True */ 00384 00385 getFloat(infile,"can_Intvl=",&can_Intvl); 00386 sprintf(msgStr,"can_Intvl=%f\n",can_Intvl); WriteMsg(msgStr,1); 00387 if ( can_Intvl == 0 ) canalCalendar = True; /* true/false flag signifying use of gregorian calendar for recurring canal (watmanage) calcs 00388 a can_Intvl of 0 days defaults to gregorian calendar-month 00389 intervals ( = #days/month, diff among months/years) 00390 In this case, the true/false can_Intvl flag is set to True */ 00391 00392 getInt(infile,"seed=",&seed); 00393 sprintf(msgStr,"seed=%d\n",seed); WriteMsg(msgStr,1); 00394 getFloat(infile,"dt=",&dt); 00395 sprintf(msgStr,"dt=%f\n",(double)dt); WriteMsg(msgStr,1); 00396 getInt(infile,"hyd_iter=",&hyd_iter); 00397 sprintf(msgStr,"hyd_iter=%d\n",hyd_iter); WriteMsg(msgStr,1); 00398 00399 sfstep = dt/hyd_iter; /* time step for cell-cell overland flows */ 00400 gwstep = dt/(hyd_iter/2); /* time step for cell-cell ground water flows (twice as long as surface water)*/ 00401 canstep = dt/hyd_iter; /* time step for canal flows */ 00402 step_Cell = sq_celWid * sfstep/CELL_SIZE * sec_per_day; /* constant used in horizontal surface water raster flux equations */ 00403 00404 N_iter= (Jdate_end - Jdate_init + 1)/dt; 00405 sprintf(msgStr,"N_iter=%d\n",N_iter); WriteMsg(msgStr,1); 00406 PORnumday = (int)(Jdate_end - Jdate_init + 1); /* number of days of simulation Period Of Record */ 00407 00408 getInt(infile,"debug=",&debug); 00409 sprintf(msgStr,"debug=%d\n",debug); WriteMsg(msgStr,1); 00410 getInt(infile,"debug_point=",&dbgPt.x); 00411 getInt(infile,NULL,&dbgPt.y); 00412 sprintf(msgStr,"debug point= (%d,%d)\n",dbgPt.x,dbgPt.y); WriteMsg(msgStr,1); 00413 00414 alloc_mem_runs(SParmNum_TEMP); 00415 RunList[0] = ProgExec; /* nominal parameter set */ 00416 00417 getString(infile,"S_ParmName=",ProgExec->S_ParmName); /* value can be: 00418 1) "NONE", for no sensitivity analysis (nominal parameter set); 00419 2) the name of a global or habitat-specific parameter for a single-parm sensitivity analysis; 00420 3) "ALL", leading to multi-parameter sensitivity analyses on 00421 all parameters, to be read from an input list */ 00422 sprintf(msgStr,"S_ParmName=%s\n",ProgExec->S_ParmName); WriteMsg(msgStr,1); 00423 00424 SensiOn = (strcmp(ProgExec->S_ParmName,"NONE")!=0) ? (True) : (False); 00425 ProgExec->S_ParmRelVal = 0; /* first run of program is ALWAYS the nominal (no parameter-changes) run */ 00426 00427 getInt(infile,"HabSwitchOn=",&HabSwitchOn); /* flag to turn on habitat switching */ 00428 sprintf(msgStr,"HabSwitchOn=%d\n",HabSwitchOn); WriteMsg(msgStr,1); 00429 getInt(infile,"WatMgmtOn=",&WatMgmtOn); /* flag to turn on canal/water management */ 00430 sprintf(msgStr,"WatMgmtOn=%d\n",WatMgmtOn); WriteMsg(msgStr,1); 00431 00432 getString(infile,"Scenario=",SimAlt); /* simulation's scenario/alternative */ 00433 sprintf(msgStr,"SimAlt=%s\n",SimAlt); WriteMsg(msgStr,1); 00434 getString(infile,"Scenario modifier=",SimModif); /* simulation's scenario/alternative & modifier */ 00435 sprintf(msgStr,"SimModif=%s\n",SimModif); WriteMsg(msgStr,1); 00436 00437 scan_forward(infile,"Sectors="); i=0; stop=0; 00438 while(1) { 00439 skip_white(infile); 00440 ch=fgetc(infile); 00441 if( isdigit(ch) ) ungetc(ch,infile); 00442 else { 00443 switch (ch) { 00444 case ';': NSector = i; stop=1; break; 00445 case ',': skip_white(infile); break; 00446 } 00447 } 00448 if(stop) break; 00449 else fscanf(infile,"%d",&iSector[i++]); 00450 /* sector 13 is the ESP (Everglades Settling-of Phosphorus, i.e., EWQModel emulation) mode - this sets the ESPmodeON flag*/ 00451 if (iSector[i-1] == 13) ESPmodeON=1; 00452 00453 } 00454 fclose(infile); 00455 broadcastInt(&NSector); 00456 sprintf(msgStr,"\n(%d) NSector = %d: Sectors=",procnum,NSector); WriteMsg(msgStr,1); 00457 for(i=0; i<NSector; i++) { 00458 broadcastInt(&iSector[i]); 00459 sprintf(msgStr,"(%d:%d)",i,iSector[i]); WriteMsg(msgStr,1); 00460 } 00461 00462 for(i=0; i<MAX_PTSERIES; i++) if(pSeries[i].data) { free((char*)pSeries[i].data); pSeries[i].data = NULL; } 00463 }
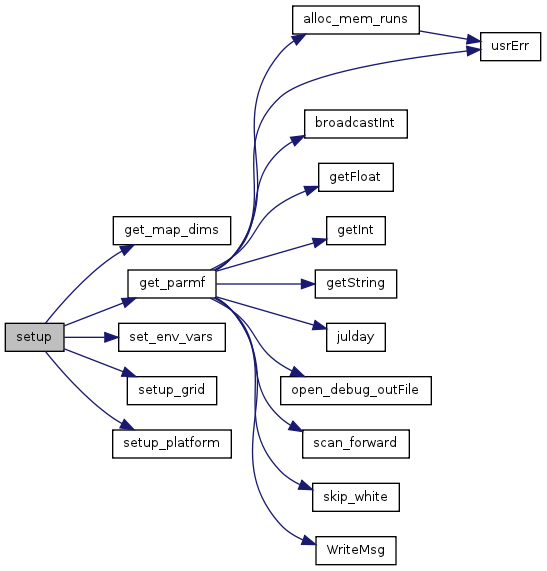
| void setup | ( | ) |
Controller to call functions to set up & initialize the model.
Several function calls here effectively do nothing in serial (i.e., non-parallel) implementations.
Definition at line 269 of file Generic_Driver.c.
References get_map_dims(), get_parmf(), MAX_PTSERIES, pSeries, set_env_vars(), setup_grid(), and setup_platform().
Referenced by main().
00270 { 00271 int i; 00272 for(i=0; i<MAX_PTSERIES; i++) pSeries[i].data = NULL; /* initialize Point Time Series arrays */ 00273 setup_platform(); /* effectively unused in serial (non-parallel) implementation */ 00274 set_env_vars(); /* get necessary enviroment variables */ 00275 get_parmf(); /* get the run-time parameters */ 00276 get_map_dims(); /* get the row-column dimensions of the global model array size */ 00277 setup_grid(); /* set up a grid size used in memory allocations (other stuff for parallel) */ 00278 }
| void track_time | ( | int | istep | ) |
Keep track of simulation time.
The elapsed days, the gregorian and julian calendars, and several end-of calendar- month, year etc flags are updated with each iteration.
- Parameters:
-
istep counter for number of model time iterations
Definition at line 473 of file Generic_Driver.c.
References avg_Intvl, avgCalendar, avgPrint, BIRavg_Intvl, BIRhyd_avg_Intvl, BIRhydCalendar, budg_Intvl, budgCalendar, calcdate(), can_Intvl, canalCalendar, canPrint, simTime::da, dryEndDay, dryEndMon, dt, False, simTime::hr, simTime::IsBIRavgEnd, simTime::IsBIRhydEnd, simTime::IsBudgEnd, simTime::IsBudgFirst, simTime::IsDay0, simTime::IsDrySeasEnd, simTime::IsMonthEnd, simTime::IsSimulationEnd, simTime::IsWetSeasEnd, simTime::IsYearEnd, simTime::Jdate, Jdate_init, simTime::mi, simTime::mo, N_iter, PORnumday, simTime::se, SensiOn, SimTime, simTime::TIME, True, wetEndDay, wetEndMon, and simTime::yr.
Referenced by main().
00474 { 00475 int yr_tom[2]; /* calendar year of tomorrow's model time */ 00476 int mo_tom[2]; /* calendar month of tomorrow's model time */ 00477 int da_tom[2]; /* calendar day of tomorrow's model time */ 00478 int hr_tom[2]; /* calendar hour of tomorrow's model time (unused in model calcs) */ 00479 int mi_tom[2]; /* calendar minute of tomorrow's model time (unused in model calcs) */ 00480 double se_tom[2]; /* calendar second of tomorrow's model time (unused in model calcs) */ 00481 00482 SimTime.TIME = istep*dt; 00483 SimTime.Jdate = Jdate_init+SimTime.TIME; /* increment the julian day counter */ 00484 00485 /* use calendar functions to determine the end-of status of today's date */ 00486 calcdate( (SimTime.Jdate+1), mo_tom, da_tom, yr_tom, hr_tom, mi_tom, se_tom); /* for TOMORROW, get the calendar date info from the julian date */ 00487 calcdate( SimTime.Jdate, SimTime.mo, SimTime.da, SimTime.yr, SimTime.hr, SimTime.mi, SimTime.se); /* for TODAY, get the calendar date info from the julian date */ 00488 00489 SimTime.IsMonthEnd = ( SimTime.mo[0] != mo_tom[0] ) ? (True) : (False); /* true/false flag signifying end of a gregorian calendar month */ 00490 SimTime.IsYearEnd = ( SimTime.yr[0] != yr_tom[0] ) ? (True) : (False); /* true/false flag signifying end of a gregorian calendar year */ 00491 SimTime.IsWetSeasEnd = (( SimTime.mo[0] == wetEndMon ) && (SimTime.da[0] == wetEndDay)) ? (True) : (False); /* true/false flag signifying end of a gregorian calendar-based wet season */ 00492 SimTime.IsDrySeasEnd = (( SimTime.mo[0] == dryEndMon ) && (SimTime.da[0] == dryEndDay)) ? (True) : (False); /* true/false flag signifying end of a gregorian calendar-based dry season */ 00493 SimTime.IsSimulationEnd = ((PORnumday-1)==istep) ? (True) : (False); 00494 00495 /* determine status of budget calc/printing flag (interval based on gregorian calendar or constant julian-day) */ 00496 if (!budgCalendar) { 00497 if ( fmod(SimTime.TIME, budg_Intvl) ==0) { 00498 /* not using gregorian calendar: at end of (constant) number of days, model time is end of budget interval */ 00499 SimTime.IsBudgEnd = True; 00500 } 00501 else { SimTime.IsBudgEnd = False; } 00502 } 00503 else { 00504 if ( SimTime.IsMonthEnd ) { 00505 /* using gregorian calendar: at calendar-month's end, model time is end of budget interval */ 00506 SimTime.IsBudgEnd = True; 00507 budg_Intvl= (float)SimTime.da[0]; /* dynamic interval becomes number of days in current gregorian-calendar month */ 00508 } 00509 else { SimTime.IsBudgEnd = False; } 00510 } 00511 00512 /* v2.8 determine status of BIR hydro calc/printing flag (interval based on gregorian calendar or constant julian-day) */ 00513 if (!BIRhydCalendar) { 00514 if ( fmod(SimTime.TIME, BIRhyd_avg_Intvl) ==0) { 00515 /* not using gregorian calendar: at end of (constant) number of days, model time is end of budget interval */ 00516 SimTime.IsBIRhydEnd = True; 00517 } 00518 else { SimTime.IsBIRhydEnd = False; } 00519 } 00520 else { 00521 if ( SimTime.IsMonthEnd ) { 00522 /* using gregorian calendar: at calendar-month's end, model time is end of budget interval */ 00523 SimTime.IsBIRhydEnd = True; 00524 BIRhyd_avg_Intvl = (float)SimTime.da[0]; /* dynamic interval becomes number of days in current gregorian-calendar month */ 00525 } 00526 else { SimTime.IsBIRhydEnd = False; } 00527 } 00528 00529 /* determine status of cell-specific averages calc/printing flag (interval based on gregorian calendar or constant julian-day) */ 00530 if (!avgCalendar) { 00531 if ( fmod(SimTime.TIME, avg_Intvl) ==0) { 00532 /* not using gregorian calendar: check for time (day) to finalize and print budgets for current interval */ 00533 avgPrint = True; } 00534 else { avgPrint = False; } 00535 } 00536 else { 00537 if ( SimTime.IsMonthEnd ) { 00538 /* using gregorian calendar: at month's end, it is time (day) to finalize and print budgets for current interval */ 00539 avgPrint = True; 00540 avg_Intvl= (float)SimTime.da[0]; /* interval becomes number of days in current gregorian-calendar month */ 00541 } 00542 else { avgPrint = False; } 00543 } 00544 00545 /* determine status of canal data calc/printing flag (interval based on gregorian calendar or constant julian-day) */ 00546 if (!canalCalendar) { 00547 if ( fmod(SimTime.TIME, can_Intvl) ==0) { 00548 /* not using gregorian calendar: check for time (day) to finalize and print canal data for current interval */ 00549 canPrint = True; } 00550 else { canPrint = False; } 00551 } 00552 else { 00553 if ( SimTime.IsMonthEnd ) { 00554 /* using gregorian calendar: at month's end, it is time (day) to finalize and print canal data for current interval */ 00555 canPrint = True; 00556 can_Intvl= (float)SimTime.da[0]; /* interval becomes number of days in current gregorian-calendar month */ 00557 } 00558 else { canPrint = False; } 00559 } 00560 00561 SimTime.IsBudgFirst = ( (SimTime.TIME/budg_Intvl) > 1 ) ? (False) : (True); 00562 SimTime.IsDay0 = (SimTime.TIME==0) ? (True) : (False); 00563 00564 /* TODO: this is to be generalized in MultiRun.c */ 00565 if (SensiOn) { 00566 BIRavg_Intvl = N_iter - 1; 00567 SimTime.IsBIRavgEnd = ( fmod(SimTime.TIME, BIRavg_Intvl) ==0) ? (True) : (False); 00568 00569 BIRhyd_avg_Intvl = N_iter - 1; 00570 SimTime.IsBIRhydEnd = ( fmod(SimTime.TIME, BIRhyd_avg_Intvl) ==0) ? (True) : (False); 00571 00572 budg_Intvl = N_iter - 1; 00573 SimTime.IsBudgEnd = ( fmod(SimTime.TIME, budg_Intvl) ==0) ? (True) : (False); 00574 } 00575 else { 00576 BIRavg_Intvl = budg_Intvl; 00577 SimTime.IsBIRavgEnd = SimTime.IsBudgEnd; 00578 } 00579 00580 }
| int PTSL_test | ( | ) |
Test for presence of point-time series meteorological data.
Most ELM applications, and particularly the regional applications, input the binary "gridIO" spatial time series data for rain and potential ET. For some fine- scaled applications, the gridIO data may be too coarse, or there may be a desire for longer simulation periods than available in the gridIO data. In those cases, if the data file named "rain.pts" (which defines the grid locations of associated time series data) exists in the application's Data directory, the simulation will attempt to use those point data for generating rainfall maps on the fly. If that is true, the "pET.pts" will also be assumed to exist (to generate potential ET maps on the fly). Finally, for these applications that generate interpolated meteorological maps on the fly, we currently (v2.6) do not use the gridIO data output from the SFWMM for depths along the ELM domain border. A more flexible data-approach may be encoded if/when needed in the future.
New function in v2.6
Definition at line 599 of file Generic_Driver.c.
References ModelPath, and ProjName.
Referenced by main().
00600 { 00601 FILE *testfile; 00602 char filename[120], ss[120]; 00603 00604 sprintf(filename,"%s/%s/Data/rain.pts",ModelPath,ProjName); 00605 testfile = fopen(filename,"r"); 00606 if(testfile==NULL) 00607 return 0; /* the point data file does not exist, will use gridIO data */ 00608 else 00609 { fclose(testfile); return 1; } /* the point data file exists, will use on-the-fly interpolations of point data */ 00610 }
| int ModExperim_test | ( | ) |
Test for presence of special parameter file for model experiments.
Most ELM applications, and particularly the regional applications, use the standard (GlobalParms and HabParms) parameter files. In special model research experiments, another parameter file may be used for specific questions. In v2.6, this is only used to modify stage/depth boundary conditions for fine-scale, long-term subregional applications.
New function in v2.6
Definition at line 622 of file Generic_Driver.c.
References ModelPath, and ProjName.
Referenced by main().
00623 { 00624 FILE *testfile; 00625 char filename[120], ss[120]; 00626 00627 sprintf(filename,"%s/%s/Data/ModExperimParms_NOM",ModelPath,ProjName); 00628 testfile = fopen(filename,"r"); 00629 if(testfile==NULL) 00630 return 0; /* the point data file does not exist, will not use any special parameters */ 00631 else 00632 { fclose(testfile); return 1; } /* the model experiment parameter file exists, will use special parameters */ 00633 }
| ViewParm* read_output_parms | ( | ) |
Call the read_output_parms function.
- Returns:
- pointer to struct of ViewParm
Definition at line 66 of file Driver_Utilities.c.
Referenced by main().
00066 { 00067 /* need to change the Outlist_size (#defined in header) if adding outlist variables to the data struct in gen_output of UnitMod.c */ 00068 ViewParm* v = readOutlist(Outlist_size); 00069 return v; 00070 }
| void read_model_parameters | ( | char * | s_parm_name, | |
| int | s_parm_relval | |||
| ) |
Call functions to read model parameters from datafiles.
- Parameters:
-
s_parm_name Name of the sensitivity parameter being varied for this run s_parm_relval Indicator of current value of relative range in sensitivity parm values: nominal (0), low (1), or high (2)
Definition at line 39 of file Driver_Utilities.c.
Referenced by main().
00039 { 00040 00041 sprintf(msgStr,"Reading global parameters..."); 00042 WriteMsg(msgStr,1); 00043 usrErr0(msgStr); 00044 00045 ReadGlobalParms(s_parm_name, s_parm_relval); 00046 sprintf(msgStr,"done."); 00047 WriteMsg(msgStr,1); 00048 usrErr(msgStr); 00049 00050 sprintf(msgStr,"Reading habitat-specific parameters..."); 00051 WriteMsg(msgStr,1); 00052 usrErr0(msgStr); 00053 00054 ReadHabParms(s_parm_name, s_parm_relval); 00055 sprintf(msgStr,"done."); 00056 WriteMsg(msgStr,1); 00057 usrErr(msgStr); 00058 00059 /* v2.6 - don't write message to console until later (when determine if parm file is used or not) */ 00060 ReadModExperimParms(s_parm_name, s_parm_relval); 00061 00062 }
| void getInt | ( | FILE * | inFile, | |
| const char * | lString, | |||
| int * | iValPtr | |||
| ) |
Get an integer following a specific string.
- Parameters:
-
inFile File that is open lString The string being read iValPtr The char value found
Definition at line 1740 of file Driver_Utilities.c.
Referenced by get_parmf().
01741 { 01742 int test; 01743 UCHAR iEx=0; 01744 if(lString) 01745 scan_forward(inFile,lString); 01746 test = fscanf(inFile,"%d",iValPtr); 01747 if(test != 1) { 01748 fprintf(stderr,"Read Error (%d):",test); 01749 if(lString) fprintf(stderr,"%s\n",lString); 01750 *iValPtr = 0; 01751 iEx = 1; 01752 } 01753 if (iEx) exit(0); 01754 }
| void getFloat | ( | FILE * | inFile, | |
| const char * | lString, | |||
| float * | fValPtr | |||
| ) |
Get a float value following a specific string.
- Parameters:
-
inFile File that is open lString The string being read fValPtr The float number found
Definition at line 1683 of file Driver_Utilities.c.
Referenced by get_parmf().
01684 { 01685 int test, fSize; 01686 UCHAR iEx=0; 01687 if(lString) scan_forward(inFile,lString); 01688 test = fscanf(inFile,"%f",fValPtr); 01689 if(test != 1) { 01690 fprintf(stderr,"Read Error (%d):",test); 01691 if(lString) fprintf(stderr,"%s\n",lString); 01692 *fValPtr = 0.0; 01693 iEx = 1; 01694 } 01695 if (iEx) exit(0); 01696 }
| void getString | ( | FILE * | inFile, | |
| const char * | lString, | |||
| char * | inString | |||
| ) |
Get a string following a specific string.
- Parameters:
-
inFile File that is open lString The string being read inString The string found
Definition at line 1722 of file Driver_Utilities.c.
Referenced by get_parmf().
01723 { 01724 int test; 01725 UCHAR iEx=0; 01726 if(lString) scan_forward(inFile,lString); 01727 test = fscanf(inFile,"%s",inString); 01728 if(test != 1) { 01729 fprintf(stderr,"Read Error (%d):",test); 01730 if(lString) fprintf(stderr,"%s\n",lString); 01731 iEx = 1; 01732 } 01733 if (iEx) exit(0); 01734 }
| void calcdate | ( | double | jd, | |
| int * | m, | |||
| int * | d, | |||
| int * | y, | |||
| int * | h, | |||
| int * | mi, | |||
| double * | sec | |||
| ) |
Determine the Gregorian date from a Julian calendar day.
Julian date converter. Takes a julian date (the number of days since some distant epoch or other), and returns an int pointer to static space. ip[0] = month; ip[1] = day of month; ip[2] = year (actual year, like 1977, not 77 unless it was 77 a.d.); ip[3] = day of week (0->Sunday to 6->Saturday) These are Gregorian. Copied from Algorithm 199 in Collected algorithms of the CACM Author: Robert G. Tantzen, Translator: Nat Howard. All taken (unmodified) from SFWMD HSM's /vol/hsm/src/libs/xmgr_julday/ directory.
- Parameters:
-
jd Julian day m Month d Day y Year h Hour mi Minute sec Second
Definition at line 1630 of file Driver_Utilities.c.
Referenced by send_point_lists2(), and track_time().
01631 { 01632 static int ret[4]; 01633 01634 long j = (long)jd; 01635 double tmp, frac = jd - j; 01636 01637 if (frac >= 0.5) { 01638 frac = frac - 0.5; 01639 j++; 01640 } 01641 else { 01642 frac = frac + 0.5; 01643 } 01644 01645 ret[3] = (j + 1L) % 7L; 01646 j -= 1721119L; 01647 *y = (4L * j - 1L) / 146097L; 01648 j = 4L * j - 1L - 146097L * *y; 01649 *d = j / 4L; 01650 j = (4L * *d + 3L) / 1461L; 01651 *d = 4L * *d + 3L - 1461L * j; 01652 *d = (*d + 4L) / 4L; 01653 *m = (5L * *d - 3L) / 153L; 01654 *d = 5L * *d - 3 - 153L * *m; 01655 *d = (*d + 5L) / 5L; 01656 *y = 100L * *y + j; 01657 if (*m < 10) 01658 *m += 3; 01659 else { 01660 *m -= 9; 01661 *y += 1; 01662 } 01663 tmp = 3600.0 * (frac * 24.0); 01664 *h = (int) (tmp / 3600.0); 01665 tmp = tmp - *h * 3600.0; 01666 01667 *mi = (int) (tmp / 60.0); 01668 *sec = tmp - *mi * 60.0; 01669 }
| void setup_platform | ( | ) |
Effectively unused in serial (non-parallel) implementation.
Definition at line 1937 of file Driver_Utilities.c.
Referenced by setup().
01937 { 01938 01939 exparam(&env); 01940 procnum = env.procnum; 01941 exgridsplit(env.nprocs, 2, nprocs); 01942 if( exgridinit(2, nprocs) < 0) { usrErr("Failed to Setup Grid"); exit(0); } 01943 exgridcoord(procnum, recpnum); 01944 }
| void set_env_vars | ( | void | ) |
Acquire necessary environment variables.
Definition at line 900 of file Driver_Utilities.c.
Referenced by setup().
00900 { 00901 00902 FILE *infile; 00903 char filename[100], ch; 00904 static long start; 00905 int i, maxLen =200; 00906 00907 ModelPath = getenv("ModelPath"); 00908 ProjName = getenv("ProjName"); 00909 DriverPath = getenv("DriverPath"); 00910 OS_TYPE = getenv("OSTYPE"); /* OSTYPE not used in code, only here for informational output */ 00911 00912 sprintf(msgStr,"OS type = %s ",OS_TYPE); 00913 usrErr(msgStr); 00914 00915 sprintf(msgStr,"Project Name = %s ",ProjName); 00916 usrErr(msgStr); 00917 00918 }
| void setup_grid | ( | ) |
Provide the 2D array size for the model domain.
Definition at line 392 of file Driver_Utilities.c.
Referenced by setup().
00392 { 00393 00394 exgridsize(procnum,gbl_size,lcl_size,lcl_start); /* effectively unused in serial (non-parallel) implementation */ 00395 00396 /* s0 and s1 remain unchanged in serial (non-parallel) implementation */ 00397 s0 = lcl_size[0]; 00398 s1 = lcl_size[1]; 00399 00400 gridSize = (s0+2)*(s1+2); /* the gridSize is increased by 2 in each dimension for buffer strips around processor domain(s) (functional mainly to parallel implementation) */ 00401 00402 sprintf(msgStr,"\nGRID DATA::[ gsize: (%d, %d), lstart: (%d, %d), lend: (%d, %d), lsize: (%d, %d) ]\n", 00403 gbl_size[0], gbl_size[1], lcl_start[0], 00404 lcl_start[1], lcl_start[0]+lcl_size[0]-1, 00405 lcl_start[1]+lcl_size[1]-1, lcl_size[0], lcl_size[1] ); 00406 WriteMsg(msgStr,1); 00407 sprintf(msgStr,"\nVP DATA:: size: (%d), Variable sizes: float: %d, int: %d, long: %d, double: %d\n", 00408 sizeof(ViewParm), sizeof(float),sizeof(int) ,sizeof(long) ,sizeof(double) ); 00409 WriteMsg(msgStr,1); 00410 00411 gTempSize = gridSize*8; 00412 gTemp = (UCHAR*) nalloc(gTempSize,"gTemp"); 00413 00414 /* v2.8.2 new temp array for output - all variables' arrays are 2 rows and 2 cols larger than the actual data 00415 this is used to hold only the size of s0, s1 */ 00416 floatOut = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"floatOut"); 00417 // init_pvar(floatOut,NULL,'f',0.0); 00418 00419 /* v2.8.2 new temp array for output - netCDF output files now have origin at lower (not upper) left */ 00420 nc_dataRowRev = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"nc_dataRowRev"); 00421 init_pvar(nc_dataRowRev,NULL,'f',0.0); 00422 00423 00424 }
| int skip_white | ( | FILE * | infile | ) |
Skip white space(s) in a file.
- Parameters:
-
infile
- Returns:
- success/failure
Definition at line 1800 of file Driver_Utilities.c.
Referenced by get_parmf(), and init_config_file().
01801 { 01802 int ch; 01803 01804 while( isspace(ch=fgetc(infile)) ) {;} 01805 if(ch==EOF) return 0; 01806 ungetc(ch,infile); 01807 return 1; 01808 }
| int scan_forward | ( | FILE * | infile, | |
| const char * | tstring | |||
| ) |
Scan forward until a particular string is found.
- Parameters:
-
infile The file being read tstring The sought-after string
- Returns:
- Success/failure
Definition at line 1814 of file Driver_Utilities.c.
Referenced by get_global_parm(), get_modexperim_parm(), get_parmf(), getChar(), getFloat(), getInt(), getString(), read_header(), read_map_dims(), and readSeriesCol().
01815 { 01816 int sLen, i, cnt=0; 01817 char Input_string[100], test; 01818 01819 sLen = strlen(tstring); 01820 while( ( test = fgetc(infile) ) != EOF ) { 01821 for(i=0; i<(sLen-1); i++) 01822 Input_string[i] = Input_string[i+1]; 01823 Input_string[sLen-1] = test; 01824 Input_string[sLen] = '\0'; 01825 if(++cnt >= sLen) { 01826 test = strcmp(Input_string,tstring); 01827 if( test == 0 ) return 1; 01828 } 01829 } 01830 return(-1); 01831 }
| double julday | ( | int | mon, | |
| int | day, | |||
| int | year, | |||
| int | h, | |||
| int | mi, | |||
| double | se | |||
| ) |
Determine the Julian calendar day from a Gregorian date.
- Returns:
- julian day + hms as a real number
- Parameters:
-
mon Month day Day year Year h Hour mi Minute se Second
- Returns:
- Julian day
Definition at line 1583 of file Driver_Utilities.c.
Referenced by get_parmf(), readSeriesCol(), ReadStructFlow_PtSer(), ReadStructTP_PtSer(), and ReadStructTS_PtSer().
01584 { 01585 long m = mon, d = day, y = year; 01586 long c, ya, j; 01587 double seconds = h * 3600.0 + mi * 60 + se; 01588 01589 if (m > 2) 01590 m -= 3; 01591 else { 01592 m += 9; 01593 --y; 01594 } 01595 c = y / 100L; 01596 ya = y - (100L * c); 01597 j = (146097L * c) / 4L + (1461L * ya) / 4L + (153L * m + 2L) / 5L + d + 1721119L; 01598 if (seconds < 12 * 3600.0) { 01599 j--; 01600 seconds += 12.0 * 3600.0; 01601 } 01602 else { 01603 seconds = seconds - 12.0 * 3600.0; 01604 } 01605 return (j + (seconds / 3600.0) / 24.0); 01606 }
| float FMOD | ( | float | x, | |
| float | y | |||
| ) |
Modulus of a pair of (double) arguments.
- Parameters:
-
x Numerator y Denominator
- Returns:
- modulus (float) result
Definition at line 1675 of file Driver_Utilities.c.
Referenced by cell_dyn1(), CellAvg(), Flows_in_Structures(), gen_output(), init_eqns(), main(), and Run_Canal_Network().
| void local_setup | ( | int | argc, | |
| char ** | argv | |||
| ) |
Does very little in serial implementation (opens a low-level debug file).
Primarily intended for parallel apps:
establish processor attributes, open a low-level debug file (in the user's current directory, set by the model execution script ("go"), and seed the pseudo-random number.
- Parameters:
-
argc number of command line arguments argv command line argument
- Returns:
- void
Definition at line 1403 of file Serial.c.
Referenced by main().
01404 { 01405 int i; 01406 char debugfileName[300]; 01407 nprocs[0] = 1; 01408 nprocs[1] = 1; 01409 tramType = 0; 01410 procnum = 1; 01411 recpnum[0] = 0; 01412 recpnum[1] = 0; 01413 Lprocnum = 1; 01414 /* the combo_table (ctable) used in Combine operation for spatial array summaries (debug-related) */ 01415 max_combos_open = 0; 01416 for(i = 0; i < MAXCOMBOS; i++) { 01417 ctable[i].ocnt = 0; 01418 ctable[i].free = 1; 01419 } 01420 01421 /* this debug output file is not used extensively in current code - it is opened before we get 01422 environment vars, thus is written to user's current directory ($ModelPath/$ProjName/Load if 01423 ELM is executed from the "go" script) */ 01424 dFile = fopen("ELM.debug","w"); 01425 if(dFile == NULL) { 01426 usrErr("Can't open ELM.debug file."); 01427 exit(0); 01428 } 01429 /* there are no random processes in current ELM (v2.4) w/o fire */ 01430 srand(seed); 01431 fprintf(dFile," RAND_MAX = %d\n",(int)RAND_MAX); 01432 }
| void open_debug_outFile | ( | int | index | ) |
Open debug-related output files.
The Driver0.out and Driver1.out are two output files with debug and/or standard error-warning information. They are always produced, but have varying levels of detail (esp. Driver1.out) depending on the user-selected (at run-time) level of the "debug" variable. Driver0.out basically provides initial, static information on data and setup of the model. Driver1.out provides dynamic information on the simulation. In sensitivity analysis mode, subsequent Driver2.out, Driver3.out, etc provide dynamic information on each new simulation.
- Parameters:
-
index Index to indicate file name w/ "0", "1", and subsequent runs under a sensitivity analysis
Definition at line 1172 of file Serial.c.
Referenced by get_parmf(), and main().
01173 { 01174 char filename[120]; 01175 01176 sprintf(filename,"%s/%s/Output/Debug/Driver%d.out",OutputPath,ProjName,index); 01177 01178 Driver_outfile = fopen(filename,"w"); 01179 if(Driver_outfile == NULL) { 01180 fprintf(stderr,"Error, unable to open %s file.\n",filename); 01181 fprintf(stderr,"OutputPath: %s\n",OutputPath); 01182 fprintf(stderr,"Project: %s\n",ProjName); 01183 exit(0); 01184 } 01185 01186 }
| void broadcastInt | ( | int * | iValPtr | ) |
Parallel code: does nothing in serial implementation).
Definition at line 1512 of file Serial.c.
Referenced by get_parmf(), PTSL_ReadLists(), and read_map_dims().
| void sync_processors | ( | ) |
| void send_point_lists2 | ( | SeriesParm * | pSeries, | |
| int | nSeries | |||
| ) |
Point time series output: print time series data to file(s).
open_point_lists Has documentation relevant to this function
- Parameters:
-
pSeries A struct of the variables to have point time series output nSeries Number of point time series occurances
Definition at line 924 of file Serial.c.
Referenced by main().
00925 { 00926 FILE *oFile; 00927 int i=0, ii, j, ix, iy, k= 0, last_pt; 00928 int yr_pt[2],mo_pt[2],da_pt[2],hr_pt[2],mi_pt[2]; 00929 double se_pt[2]; 00930 double Jdate_pt; 00931 00932 if (numPtFiles==0) return; 00933 /* k = file/variable counter; i = indiv cellLoc time series counter */ 00934 for (k = 0; k <= numPtFiles; k++) { /* loop over the number of point ts files */ 00935 sprintf(msgStr,"%s%s/Output/PtSer/%s.pts",OutputPath,ProjName,pSeries[i].name); 00936 if( (oFile = fopen(msgStr,"a") ) == NULL) { 00937 fprintf(stderr,"\nERROR, unable to open %s point time series output file.",msgStr); 00938 exit(-1); 00939 } 00940 00941 for(j=pSeries[i].laststep; j<pSeries[i].Length; j++ ) { /*temporal loop */ 00942 Jdate_pt = Jdate_init+j*pSeries[i].outstep; 00943 calcdate( Jdate_pt, mo_pt, da_pt, yr_pt, hr_pt, mi_pt, se_pt); /* get the calendar date info from the julian date */ 00944 fprintf(oFile,"\n%d/%d/%d\t",yr_pt[0],mo_pt[0],da_pt[0] ); /* calendar date in col 1 */ 00945 00946 last_pt = i+cell_pts[k]; /* # of last cellLoc point of the current variable's file */ 00947 for (ii=i; ii<last_pt; ii++) {/* loop over number of point locations per file */ 00948 fprintf(oFile,"%f\t",pSeries[ii].data[j]); 00949 } 00950 } 00951 fclose (oFile); 00952 pSeries[i].laststep = j; /* remember the last temporal outstep for this file */ 00953 i += cell_pts[k]; /* increment the indiv cellLoc time series counter */ 00954 } 00955 return; 00956 }
| void open_point_lists | ( | SeriesParm * | pSeries, | |
| int | nSeries | |||
| ) |
Point time series output: open files and print headers.
Provides for multi-column (locations), multi-file (model variables)format of point time series. The user can define (via Model.outList commands), for any variable in the model, one or more cell (point) locations for which point time series data are written. There can be multiple point TS each for different variables. Thus, we make a separate file for each variable (unknown until user chooses) that has multiple columns for multiple points (unknown until user chooses).
- Parameters:
-
pSeries An array of SeriesParm structs for point time series output nSeries Number of point time series occurances
- Remarks:
- A structure should/could be set up for this, but to maintain minimal changes between the parallel and serial versions of the code (ELMv1.0), for now we just maintain the original data structure and use some counters below to keep track of diff files and numbers of columns (also allowing different output steps as chosen by the user). (Note: The original SME code printed all variables in one column in one file).
- Note:
- TODO: this whole point-time-series output functionality needs improvement (not high priority, Jan 2005). One of the more important capabilities is to have ability to have irregular, calendar-month/year etc output intervals.
Definition at line 881 of file Serial.c.
Referenced by main().
00882 { 00883 FILE *oFile; 00884 int i, j, ix, iy, k= 0 ; 00885 00886 for (i=0; i<nSeries; i++) { 00887 if( pSeries[i].data == NULL ) { 00888 if ( numPtFiles>0 ) fclose(oFile); /* in (rare?) case where user asks for no point output, no files stream to close */ 00889 return; 00890 } 00891 if (strcmp(pSeries[i].name,pSeries[i-1].name) != 0) { 00892 /* this is new variable, thus new file. numPtFiles== increment counter of number of variables (one file per variable) used for output */ 00893 if (i>0) { 00894 numPtFiles++; 00895 fclose (oFile); 00896 } 00897 00898 sprintf(msgStr,"%s%s/Output/PtSer/%s.pts",OutputPath,ProjName,pSeries[i].name); 00899 if( (oFile = fopen(msgStr,"w") ) == NULL) { 00900 fprintf(stderr,"\nERROR, unable to open %s point time series output file.",msgStr); 00901 exit(-1); 00902 } 00903 fprintf (oFile, "%s %s %s %s scenario: \nDate\t", &modelName, &modelVers, &SimAlt, &SimModif); 00904 00905 } 00906 /* populate the grid cell locations in the header for this variable/file */ 00907 ix = pSeries[i].Loc.x; 00908 iy = pSeries[i].Loc.y; 00909 fprintf(oFile,"%s:(%d,%d)\t",pSeries[i].name,ix,iy); 00910 cell_pts[numPtFiles]++; /* count the number of cell point locs per variable (1 var per file) */ 00911 00912 } 00913 }
| void init_static_data | ( | void | ) |
Initialize static spatial data.
Definition at line 1710 of file UnitMod.c.
Referenced by main().
01711 { 01712 usrErr0("Acquiring static spatial data..."); /* console message */ 01713 01714 read_map_file("ModArea",ON_MAP,'c',4.0,0.0); /* defines model area, dimless unsigned char attributes, value 1 set to 4, 0=0 */ 01715 read_map_file("BoundCond",BCondFlow,'d',1.0,0.0); /* boundary condition flow restrictions, dimless integer attributes */ 01716 /* ONLY when running the EWQM settling rate version of ELM */ 01717 if (ESPmodeON) read_map_file("basinSetVel",WQMsettlVel,'f',0.0001,0.0); /* basin-based settling rates (data in tenths mm/d, converted to m/d) */ 01718 read_map_file("basins",basn,'d',1.0,0.0); /* Basins/Indicator-Region map, dimless integer attributes */ 01719 01720 alloc_mem_stats (); /* allocate memory for budget & stats arrays (need to have read the Basin/Indicator-Region map first) */ 01721 BIRinit(); /* Set up the Basin/Indicator-Region (BIR) linkages/inheritances */ 01722 BIRoutfiles(); /* Basin/Indicator-Region output files */ 01723 01724 usrErr("Done."); 01725 }
| void init_dynam_data | ( | void | ) |
Initialize dynamic spatial data.
Definition at line 1729 of file UnitMod.c.
Referenced by main().
01730 { 01731 usrErr0("Acquiring dynamic spatial data..."); /* console message */ 01732 01733 read_map_file("Elevation",ELEVATION,'f',0.01,0.0); /* positive elevation relative to zero of vertical datum; input data in cm, converted to m; (prior to ELMv2.8, datum was always NGVD 1929; some apps in v2.8+ are in NAVD88; datum is independent of source code ) */ 01734 read_map_file("HAB",HAB,'c',1.0,0.0); /* habitat (veg classifications, dimless unsigned char attributes) */ 01735 read_map_file("icMacBio",MAC_TOT_BIOM,'f',0.015,0.0); /* initial total biomass of macrophytes (data in xyz, converted to kg C/m2) */ 01736 read_map_file("icSfWt",SURFACE_WAT,'f',0.01,0.0); /* initial ponded surface water (data in cm, converted to m) */ 01737 read_map_file("icUnsat",UNSAT_DEPTH,'f',0.01,0.0); /* initial depth of unsaturated zone (data in cm, converted to m) */ 01738 read_map_file("soilTP",TPtoSOIL,'f',0.000001,0.0); /* soil TP map (data in mgP/kgSoil, converted to kgP/kgSoil) */ 01739 read_map_file("soilBD",BulkD,'f',1.0,0.0); /* soil bulk dens map (data in mgSoil/cm3 == kgSoil/m3) */ 01740 read_map_file("soil_orgBD",DOM_BD,'f',1.0,0.0); /* organic soil bulk dens map (data in mgOM/cm3 == kgOM/m3) */ 01741 read_map_file("soilTPpore",TP_SEDWT_CONCACT,'f',0.000001,0.0); /* porewater TP concentration map (data in ug/L, converted to g/L == kg/m3) */ 01742 read_map_file("AtmosPdepos",TP_AtmosDepos,'f',1.0,0.0); /* rate of atmospheric deposition of total phosphorus (data in mgP/m^2/yr, convert units later with eqn inits) */ 01743 01744 /* 2 static mapps need to be here for re-initialing w/ a multiplier (sensitivity analysis) */ 01745 read_map_file("HydrCond",HYD_RCCONDUCT,'f',1.0,0.0); /* hydraulic conductivity (no conversion, data in m/d) */ 01746 read_map_file("Bathymetry",Bathymetry,'f',0.01,0.0); /* positive bathymetry (depth) relative to zero of vertical datum used in land elevation surveys (i.e., positive depth below NGVD29 or NAVD88); all positive depth data in cm, converted to m) */ 01747 01748 usrErr("Done."); /* console message */ 01749 }
| void init_eqns | ( | void | ) |
Initialization of the model equations.
This calculates initial values of state variables, and which also provides constant values of pertinent flux equations if their corresponding cell_dyn module is not executed.
Definition at line 1757 of file UnitMod.c.
Referenced by main().
01758 { 01759 int ix,iy, cellLoc; 01760 float tmp; /* used in checking nutrient availability for MichMent kinetics */ 01761 float min_litTemp; /* used to determine min of temper, light cf's for alg and macs */ 01762 float I_ISat, Z_extinct, PO4Pconc, PO4P; 01763 float MAC_PHtoNPH, MAC_PHtoNPH_Init; 01764 double soil_propOrg; 01765 #define satDensity 0.9 /* assign the relative proportion (0 - 1) of a population maximum to be the saturation density for a population */ 01766 01767 usrErr0("Initializing unit model equations..."); /* console message */ 01768 01769 SimTime.TIME = 0; 01770 DAYJUL = ( FMOD(SimTime.TIME,365.0) >0.0 ) ? ( FMOD(SimTime.TIME,365.0) ) : ( 365.0); 01771 LATRAD = ((int)(GP_LATDEG)+(GP_LATDEG-(int)(GP_LATDEG))*5.0/3.0)*PI/180.0; 01772 /* AMPL = Exp(7.42+0.045*LATRAD*180.0/PI)/3600.0; */ 01773 /* DAYLENGTH = AMPL*Sin((DAYJUL-79.0)*0.01721)+12.0; */ 01774 SOLALPHA = 32.9835-64.884*(1.0-1.3614*Cos(LATRAD)); 01775 SOLDEC1 = 0.39785*Sin(4.868961+0.017203*DAYJUL +0.033446*Sin(6.224111+0.017202*DAYJUL)); 01776 SOLCOSDEC = sqrt(1.0-pow(SOLDEC1,2.0)); 01777 SOLELEV_SINE = Sin(LATRAD)*SOLDEC1+Cos(LATRAD)*SOLCOSDEC; 01778 SOLALTCORR = (1.0-Exp(-0.014*(GP_ALTIT-274.0)/(SOLELEV_SINE*274.0))); 01779 SOLBETA = 0.715-0.3183*(1.0-1.3614*Cos(LATRAD)); 01780 SOLDEC = Arctan(SOLDEC1/sqrt(1.0-pow(SOLDEC1,2.0))); 01781 SOLRISSET_HA1 = -Tan(LATRAD)*Tan(SOLDEC); 01782 SOLRISSET_HA = ( (SOLRISSET_HA1==0.0) ) ? ( PI*0.5 ) : ( ( (SOLRISSET_HA1<0.0) ) ? ( PI+Arctan(sqrt(1.0-pow(SOLRISSET_HA1,2.0))/SOLRISSET_HA1) ) : ( Arctan(sqrt(1.0-pow(SOLRISSET_HA1,2.0))/SOLRISSET_HA1))); 01783 SOLRADATMOS = 458.37*2.0*(1.0+0.033*Cos(360.0/365.0*PI/180.0*DAYJUL))* (Cos(LATRAD)*Cos(SOLDEC)*Sin(SOLRISSET_HA) + SOLRISSET_HA*180.0/(57.296*PI)*Sin(LATRAD)*Sin(SOLDEC)); 01784 01785 for(ix=0; ix<=s0+1; ix++) { 01786 for(iy=0; iy<=s1+1; iy++) { 01787 cellLoc = T(ix,iy); 01788 01789 AIR_TEMP[cellLoc] = 25.0; /* spatial time series unavailable after 1995; globally constant in v2.2+ */ 01790 /* rainfall read from sfwmm data, remapped to ELM resolution */ 01791 boundcond_rain[cellLoc] = SF_WT_FROM_RAIN[cellLoc] = boundcond_ETp[cellLoc] = boundcond_depth[cellLoc] = 0.0; 01792 01793 /* used to have cloudiness influence on GP_SOLOMEGA, now 0 */ 01794 SOLRAD274[cellLoc] = SOLRADATMOS*(SOLBETA-GP_SOLOMEGA*0.0 )-SOLALPHA; 01795 SOLRADGRD[cellLoc] = SOLRAD274[cellLoc]+((SOLRADATMOS+1.0)-SOLRAD274[cellLoc])*SOLALTCORR; 01796 H2O_TEMP[cellLoc] = AIR_TEMP[cellLoc]; 01797 01798 ALG_REFUGE[cellLoc] = HP_ALG_MAX[HAB[cellLoc]]*GP_ALG_REF_MULT; 01799 ALG_SAT[cellLoc] = HP_ALG_MAX[HAB[cellLoc]]*0.9; 01800 01801 /* v2.3: with southern everglades topo, put bathymetry back into the model */ 01802 ELEVATION[cellLoc] = ELEVATION[cellLoc] * GP_IC_ELEV_MULT; 01803 Bathymetry[cellLoc] = Bathymetry[cellLoc] * GP_IC_BATHY_MULT; 01804 SED_ELEV[cellLoc] = ELEVATION[cellLoc] - Bathymetry[cellLoc] + GP_DATUM_DISTANCE; 01805 SED_INACT_Z[cellLoc] = SED_ELEV[cellLoc]-HP_DOM_MAXDEPTH[HAB[cellLoc]]; 01806 01807 MAC_MAX_BIO[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]]+HP_PHBIO_MAX[HAB[cellLoc]]; 01808 NPHBIO_REFUGE[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]]*GP_MAC_REFUG_MULT; 01809 NPHBIO_SAT[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]] * satDensity; 01810 PHBIO_REFUGE[cellLoc] = HP_PHBIO_MAX[HAB[cellLoc]]*GP_MAC_REFUG_MULT; 01811 PHBIO_SAT[cellLoc] = HP_PHBIO_MAX[HAB[cellLoc]] * satDensity; 01812 } 01813 } 01814 for(ix=1; ix<s0+1; ix++) { 01815 for(iy=1; iy<s1+1; iy++) { cellLoc = T(ix,iy); 01816 /*initialization of major state variables */ 01817 01818 /* hydro */ 01819 HYD_RCCONDUCT[cellLoc] = HYD_RCCONDUCT[cellLoc] * GP_calibGWat; 01820 01821 /* map */ UNSAT_DEPTH[cellLoc] = Max(UNSAT_DEPTH[cellLoc] + GP_HYD_IC_UNSAT_ADD,0.0); /* m */ 01822 SAT_WT_HEAD[cellLoc] = SED_ELEV[cellLoc]- UNSAT_DEPTH[cellLoc]; /* m */ 01823 SAT_WATER[cellLoc] = SAT_WT_HEAD[cellLoc] * HP_HYD_POROSITY[HAB[cellLoc]]; /* m */ 01824 01825 UNSAT_WATER[cellLoc] = HP_HYD_POROSITY[HAB[cellLoc]]*UNSAT_DEPTH[cellLoc] *GP_HYD_ICUNSATMOIST; /* m */ 01826 UNSAT_CAP[cellLoc] = UNSAT_DEPTH[cellLoc]*HP_HYD_POROSITY[HAB[cellLoc]]; /* m */ 01827 UNSAT_MOIST_PRP[cellLoc] = /* dimless proportion, 0-1 */ 01828 ( UNSAT_CAP[cellLoc]>0.0 ) ? 01829 ( Min(UNSAT_WATER[cellLoc]/UNSAT_CAP[cellLoc],1.0) ) : 01830 ( 1.0); 01831 01832 /* map */ SURFACE_WAT[cellLoc] = Max(SURFACE_WAT[cellLoc] + GP_HYD_IC_SFWAT_ADD, 0.0); /* m */ 01833 SFWT_VOL[cellLoc] = SURFACE_WAT[cellLoc]*CELL_SIZE; 01834 01835 HYD_DOM_ACTWAT_VOL[cellLoc] = Max(HP_DOM_MAXDEPTH[HAB[cellLoc]]-UNSAT_DEPTH[cellLoc]* 01836 (1.0-UNSAT_MOIST_PRP[cellLoc]),0.0)*HP_HYD_POROSITY[HAB[cellLoc]]*CELL_SIZE; 01837 HYD_SED_WAT_VOL[cellLoc] = (SAT_WATER[cellLoc]+UNSAT_WATER[cellLoc])*CELL_SIZE; 01838 01839 /* soil */ 01840 /* map */ DOM_BD[cellLoc] = DOM_BD[cellLoc] * GP_IC_DOM_BD_MULT; /* kg/m3 */ 01841 /* map */ BulkD[cellLoc] = BulkD[cellLoc] * GP_IC_BulkD_MULT; /* kgOM/m3 */ 01842 01843 soil_propOrg = (double) DOM_BD[cellLoc] / BulkD[cellLoc]; 01844 DIM[cellLoc] = (1.0 - soil_propOrg) * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] * CELL_SIZE; /* kg inorganic matter */ 01845 DEPOS_ORG_MAT[cellLoc] = soil_propOrg * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] ; /* kg organic matter/m2 */ 01846 // DEPOS_ORG_MAT[cellLoc] = soil_propOrg * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] ; /* kg organic matter/m2 */ 01847 /* v2.8.4 new DEPOS_ORG_MAT init */ 01848 01849 // Inorg_Z[cellLoc] = (1.0 - soil_propOrg) * HP_DOM_MAXDEPTH[HAB[cellLoc]]; /* fixed inorganic depth (m) */ 01850 // DOM_Z[cellLoc] = HP_DOM_MAXDEPTH[HAB[cellLoc]] - Inorg_Z[cellLoc]; /* m */ 01851 Inorg_Z[cellLoc] = DIM[cellLoc] / BulkD[cellLoc] * (1.0-soil_propOrg) / CELL_SIZE ; /* kgInorg / kgSoil/m3 * proprInorg / m2 == fixed inorganic depth (m) */ 01852 DOM_Z[cellLoc] = HP_DOM_MAXDEPTH[HAB[cellLoc]] - Inorg_Z[cellLoc]; /* m */ 01853 01854 // v2.8.4 DEPOS_ORG_MAT[cellLoc] = DOM_BD[cellLoc]*DOM_Z[cellLoc]; /* (mgOM/cm3 ==> kgOM/m3) * m = kgOM/m2 */ 01855 01856 DOM_SED_AEROB_Z[cellLoc] = Min(UNSAT_DEPTH[cellLoc]+HP_DOM_AEROBTHIN[HAB[cellLoc]],HP_DOM_MAXDEPTH[HAB[cellLoc]]); /* m */ 01857 DOM_SED_ANAEROB_Z[cellLoc] = HP_DOM_MAXDEPTH[HAB[cellLoc]]-DOM_SED_AEROB_Z[cellLoc]; /* m */ 01858 01859 /* map */ TPtoSOIL[cellLoc] = TPtoSOIL[cellLoc] * GP_IC_TPtoSOIL_MULT; /* kgP/kgSoil */ 01860 DOP[cellLoc] = (double) (1.0-GP_sorbToTP) * TPtoSOIL[cellLoc] * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] ; /* kgP/kg_soil * kg_soil/m3 * m == kgP/m2 */ 01861 01862 TP_SORB[cellLoc] = (double) GP_sorbToTP * TPtoSOIL[cellLoc] * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] * CELL_SIZE; /* dimless * kgP/kg_soil * kg_soil/m3 * m * m^2 == kgP */ 01863 01864 /* floc layer of soil */ 01865 FLOC[cellLoc] = HP_FLOC_IC[HAB[cellLoc]]; /* kg OM/m2 */ 01866 FlocP[cellLoc] = FLOC[cellLoc]*HP_FLOC_IC_PC[HAB[cellLoc]]*HP_FLOC_IC_CTOOM[HAB[cellLoc]]; /* kg P /m2 */ 01867 FLOC_Z[cellLoc] = (double) FLOC[cellLoc] / GP_Floc_BD ; /* m */ 01868 01869 /* phosphorus */ 01870 TP_AtmosDepos[cellLoc] = (double)TP_AtmosDepos[cellLoc] * conv_mgTOkg * CELL_SIZE / 365.0; /* P deposition, mgP/m2/yr * kg/mg * m2 / daysPerYear = kgP/day */ 01871 /* v2.4.4 */ TP_SFWT_CONC[cellLoc] = GP_TP_ICSFWAT * conv_mgTOg; /* mg/L * g/mg => g/L */ 01872 TP_SF_WT[cellLoc] =TP_SFWT_CONC[cellLoc] * SFWT_VOL[cellLoc]; /* kg P */ 01873 TP_SFWT_CONC_MG[cellLoc] = TP_SFWT_CONC[cellLoc] * conv_gTOmg; /* mg/L */ 01874 /* using regression for predicting PO4 from TP */ 01875 PO4Pconc = Max(TP_SFWT_CONC_MG[cellLoc]*GP_PO4toTP + GP_PO4toTPint, 0.10 * TP_SFWT_CONC_MG[cellLoc]); /* mg/L */ 01876 PO4P = PO4Pconc * SFWT_VOL[cellLoc] /conv_kgTOg; /*kg P available (from conc. in mg/l) */ 01877 01878 /* v2.4.4 */ // TP_SED_CONC[cellLoc] = GP_TP_ICSEDWAT * conv_mgTOg; /* mg/L * g/mg => g/L */ 01879 // TP_SED_WT[cellLoc] = TP_SED_CONC[cellLoc] * HYD_SED_WAT_VOL[cellLoc]; /* kg P */ 01880 // /* this is the active zone, where uptake, sorption, and mineralization take place */ 01881 // TP_Act_to_Tot[cellLoc] = 1.0 / HP_TP_CONC_GRAD[HAB[cellLoc]]; /* ratio of TP conc in active zone relative to total */ 01882 // TP_SED_WT_AZ[cellLoc] = TP_SED_CONC[cellLoc] * TP_Act_to_Tot[cellLoc] * HYD_DOM_ACTWAT_VOL[cellLoc]; /* kg P */ 01883 // TP_SEDWT_CONCACT[cellLoc] =(HYD_DOM_ACTWAT_VOL[cellLoc] > 0.0) ? 01884 // ( TP_SED_WT_AZ[cellLoc]/HYD_DOM_ACTWAT_VOL[cellLoc] ) : 01885 // ( TP_SED_CONC[cellLoc]); /* g/L */ 01886 // TP_SEDWT_CONCACTMG[cellLoc] = TP_SEDWT_CONCACT[cellLoc]*conv_gTOmg; /* mg/L */ 01887 01888 /* v2.8.4 concentration in Active Zone (TP_SEDWT_CONCACT) is initialed by map (data here are g/L) */ 01889 TP_Act_to_Tot[cellLoc] = 1.0 / HP_TP_CONC_GRAD[HAB[cellLoc]]; /* ratio of TP conc in active zone relative to total */ 01890 TP_SED_CONC[cellLoc] = (TP_Act_to_Tot[cellLoc]>0.0) ? (TP_SEDWT_CONCACT[cellLoc] / TP_Act_to_Tot[cellLoc]) : (0.0); 01891 /* map */ TP_SEDWT_CONCACT[cellLoc] =(HYD_DOM_ACTWAT_VOL[cellLoc] > 0.0) ? 01892 ( TP_SEDWT_CONCACT[cellLoc] ) : 01893 ( 0.0 ); /* g/L */ 01894 TP_SEDWT_CONCACTMG[cellLoc] = TP_SEDWT_CONCACT[cellLoc]*conv_gTOmg; /* mg/L */ 01895 TP_SED_WT[cellLoc] = TP_SED_CONC[cellLoc] * HYD_SED_WAT_VOL[cellLoc]; /* kg P */ 01896 TP_SED_WT_AZ[cellLoc] = TP_SEDWT_CONCACT[cellLoc] * HYD_DOM_ACTWAT_VOL[cellLoc]; /* kg P */ 01897 01898 01899 /* salt */ 01900 SALT_SED_WT[cellLoc] = HYD_SED_WAT_VOL[cellLoc]*HP_SALT_ICSEDWAT[HAB[cellLoc]]; 01901 SAL_SED_WT[cellLoc] = ( HYD_SED_WAT_VOL[cellLoc]>0.0 ) ? ( SALT_SED_WT[cellLoc]/HYD_SED_WAT_VOL[cellLoc] ) : ( 0.0); 01902 SALT_SURF_WT[cellLoc] = SFWT_VOL[cellLoc]*HP_SALT_ICSFWAT[HAB[cellLoc]]; 01903 SAL_SF_WT[cellLoc] = ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? ( SALT_SURF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : ( 0.0); 01904 01905 /* periphyton */ 01906 /* 2.4.4 */ NC_ALG[cellLoc] = HP_ALG_MAX[HAB[cellLoc]] * GP_ALG_IC_MULT * GP_ALG_REF_MULT ; /* start w/ low, refuge-level of non-calcareous (eutrophic) periphyton, g C/m2 */ 01907 /* 2.4.4 */ C_ALG[cellLoc] = HP_ALG_MAX[HAB[cellLoc]] * GP_ALG_IC_MULT * (1.0 - GP_ALG_REF_MULT); /* g C/m2 */ 01908 /* ic PC of periph in oligotrophic area is 3% of max P:C, varies across space via (0.1->1.0) map */ 01909 /* 2.4.4 */ NC_ALG_PC[cellLoc] = GP_ALG_PC; /* gP/ gC */ 01910 /* 2.4.4 */ C_ALG_PC[cellLoc] = GP_ALG_PC; /* gP/ gC */ 01911 01912 NC_ALG_P[cellLoc] = NC_ALG[cellLoc]*NC_ALG_PC[cellLoc]; /* g P/m2 */ 01913 C_ALG_P[cellLoc] = C_ALG[cellLoc]*C_ALG_PC[cellLoc]; /* g P/m2 */ 01914 01915 /* macrophytes */ 01916 /* 2.4.4 */ MAC_PH_BIOMAS[cellLoc] = MAC_TOT_BIOM[cellLoc] * GP_MAC_IC_MULT * HP_PHBIO_MAX[HAB[cellLoc]]/MAC_MAX_BIO[cellLoc]; /* kg C/m2 */ 01917 /* now calc the P and OM associated with that C */ 01918 /* 2.4.4 */ mac_ph_PC[cellLoc] = HP_PHBIO_IC_PC[HAB[cellLoc]]; 01919 mac_ph_P[cellLoc] = MAC_PH_BIOMAS[cellLoc] * mac_ph_PC[cellLoc]; /* kg P/m2 */ 01920 mac_ph_OM[cellLoc] = MAC_PH_BIOMAS[cellLoc] / HP_PHBIO_IC_CTOOM[HAB[cellLoc]]; 01921 mac_ph_CtoOM[cellLoc] = HP_PHBIO_IC_CTOOM[HAB[cellLoc]]; 01922 PHBIO_AVAIL[cellLoc] = MAC_PH_BIOMAS[cellLoc]*Max(1.0-Max((PHBIO_SAT[cellLoc]-MAC_PH_BIOMAS[cellLoc]) /(PHBIO_SAT[cellLoc]-PHBIO_REFUGE[cellLoc]),0.0),0.0); 01923 01924 /* 2.4.4 */ MAC_NOPH_BIOMAS[cellLoc] = MAC_TOT_BIOM[cellLoc] * GP_MAC_IC_MULT * HP_NPHBIO_MAX[HAB[cellLoc]]/MAC_MAX_BIO[cellLoc]; /* kg C/m2 */ 01925 /* now calc the P and OM associated with that C */ 01926 /* 2.4.4 */ mac_nph_PC[cellLoc] = HP_NPHBIO_IC_PC[HAB[cellLoc]]; 01927 mac_nph_P[cellLoc] = MAC_NOPH_BIOMAS[cellLoc] * mac_nph_PC[cellLoc]; /* kg P/m2 */ 01928 mac_nph_OM[cellLoc] = MAC_NOPH_BIOMAS[cellLoc] / HP_NPHBIO_IC_CTOOM[HAB[cellLoc]]; 01929 mac_nph_CtoOM[cellLoc] = HP_NPHBIO_IC_CTOOM[HAB[cellLoc]]; 01930 NPHBIO_AVAIL[cellLoc] = MAC_NOPH_BIOMAS[cellLoc]*Max(1.0-Max((NPHBIO_SAT[cellLoc]-MAC_NOPH_BIOMAS[cellLoc])/(NPHBIO_SAT[cellLoc]-NPHBIO_REFUGE[cellLoc]),0.0),0.0); 01931 01932 MAC_REL_BIOM[cellLoc] = ( MAC_TOT_BIOM[cellLoc] > 0 ) ? MAC_TOT_BIOM[cellLoc]/MAC_MAX_BIO[cellLoc] : 0.0; 01933 MAC_LAI[cellLoc] = MAC_REL_BIOM[cellLoc]*HP_MAC_MAXLAI[HAB[cellLoc]]; 01934 MAC_HEIGHT[cellLoc] = MAC_REL_BIOM[cellLoc]*HP_MAC_MAXHT[HAB[cellLoc]]; 01935 LAI_eff[cellLoc] = (MAC_HEIGHT[cellLoc]>0.0) ? (Max(1.0 - SURFACE_WAT[cellLoc]/MAC_HEIGHT[cellLoc], 0.0)*MAC_LAI[cellLoc]) : (0.0) ; 01936 01937 /* end of initialization of major state variables */ 01938 01939 /* *************************** */ 01940 /* These are the multiple calculations used if particular modules are turned off. \n 01941 NOTE: THIS section (of init_eqns() ) is not fully updated for properly 01942 turning off individual **interacting** *biological/chemical* modules. 01943 If one *biological/chemical* module is turned off, 01944 they all need to be turned off. (Note that cell_dyn's 3,5,6 should always be off). \n 01945 01946 *** \n 01947 The following *biological/chemical* modules must be ON or OFF as a group: 01948 (cell_dyn2 + cell_dyn4 + cell_dyn8 + cell_dyn9 + cell_dyn12) 01949 cell_dyn13, the net settling rate module, can be turned on only when above module group is off 01950 *** \n 01951 01952 In particular, the budget will 01953 not properly reflect actual dynamics if those 01954 modules are not treated as a group. 01955 */ 01956 NC_ALG_MORT_POT[cellLoc] = ( UNSAT_DEPTH[cellLoc]>0.05 ) ? ( NC_ALG[cellLoc]*GP_ALG_RC_MORT_DRY ) : ( NC_ALG[cellLoc]*GP_ALG_RC_MORT); 01957 /* calcareous periphyton */ 01958 C_ALG_MORT_POT[cellLoc] = ( UNSAT_DEPTH[cellLoc]>0.05 ) ? ( C_ALG[cellLoc]*GP_ALG_RC_MORT_DRY ) : ( C_ALG[cellLoc]*GP_ALG_RC_MORT); 01959 ALG_TEMP_CF[cellLoc] = tempCF(0, 0.20, GP_ALG_TEMP_OPT, 5.0, 40.0, H2O_TEMP[cellLoc]); 01960 NC_ALG_RESP_POT[cellLoc] = 01961 ( UNSAT_DEPTH[cellLoc]<0.05 ) ? 01962 ( GP_ALG_RC_RESP*ALG_TEMP_CF[cellLoc]*NC_ALG[cellLoc] ) : 01963 ( 0.0); 01964 NC_ALG_RESP[cellLoc] = 01965 ( NC_ALG_RESP_POT[cellLoc]*DT>NC_ALG[cellLoc] ) ? 01966 ( NC_ALG[cellLoc]/DT ) : 01967 ( NC_ALG_RESP_POT[cellLoc]); 01968 /* calcareous periphyton */ 01969 C_ALG_RESP_POT[cellLoc] = 01970 ( UNSAT_DEPTH[cellLoc]<0.05 ) ? 01971 ( GP_ALG_RC_RESP*ALG_TEMP_CF[cellLoc]*C_ALG[cellLoc] ) : 01972 ( 0.0); 01973 C_ALG_RESP[cellLoc] = 01974 ( C_ALG_RESP_POT[cellLoc]*DT>C_ALG[cellLoc] ) ? 01975 ( C_ALG[cellLoc]/DT ) : 01976 ( C_ALG_RESP_POT[cellLoc]); 01977 01978 NC_ALG_AVAIL_MORT[cellLoc] = NC_ALG[cellLoc]-ALG_REFUGE[cellLoc]; 01979 NC_ALG_MORT[cellLoc] = ( (NC_ALG_MORT_POT[cellLoc])*DT>NC_ALG_AVAIL_MORT[cellLoc] ) ? ( (NC_ALG_AVAIL_MORT[cellLoc])/DT ) : ( NC_ALG_MORT_POT[cellLoc]); 01980 /* calcareous periphyton */ 01981 C_ALG_AVAIL_MORT[cellLoc] = C_ALG[cellLoc]-ALG_REFUGE[cellLoc]; 01982 C_ALG_MORT[cellLoc] = ( (C_ALG_MORT_POT[cellLoc])*DT>C_ALG_AVAIL_MORT[cellLoc] ) ? ( (C_ALG_AVAIL_MORT[cellLoc])/DT ) : ( C_ALG_MORT_POT[cellLoc]); 01983 01984 /* light, water, temperature controls apply to calc and non-calc periphyton */ 01985 ALG_LIGHT_EXTINCT[cellLoc] = 0.01; /* light extinction coef */ 01986 /* algal self-shading implicit in density-dependent constraint function later */ 01987 ALG_INCID_LIGHT[cellLoc] = SOLRADGRD[cellLoc]*Exp(-MAC_LAI[cellLoc]*GP_ALG_SHADE_FACTOR); 01988 Z_extinct = SURFACE_WAT[cellLoc]*ALG_LIGHT_EXTINCT[cellLoc]; 01989 I_ISat = ALG_INCID_LIGHT[cellLoc]/GP_ALG_LIGHT_SAT; 01990 /* averaged over whole water column (based on Steele '65) */ 01991 ALG_LIGHT_CF[cellLoc] = ( Z_extinct > 0.0 ) ? 01992 ( 2.718/Z_extinct * (Exp(-I_ISat * Exp(-Z_extinct)) - Exp(-I_ISat)) ) : 01993 (I_ISat*Exp(1.0-I_ISat)); 01994 /* low-water growth constraint ready for something better based on data */ 01995 ALG_WAT_CF[cellLoc] = ( SURFACE_WAT[cellLoc]>0.0 ) ? ( 1.0 ) : ( 0.0); 01996 /* the 2 communities have same growth response to avail phosphorus - avail P is roughly calc'd from TP */ 01997 NC_ALG_NUT_CF[cellLoc] = PO4Pconc/(PO4Pconc+GP_NC_ALG_KS_P) ; 01998 C_ALG_NUT_CF[cellLoc] = PO4Pconc/(PO4Pconc+GP_C_ALG_KS_P); 01999 min_litTemp = Min(ALG_LIGHT_CF[cellLoc],ALG_TEMP_CF[cellLoc]); 02000 NC_ALG_PROD_CF[cellLoc] = Min(min_litTemp,ALG_WAT_CF[cellLoc])*NC_ALG_NUT_CF[cellLoc]; 02001 C_ALG_PROD_CF[cellLoc] = Min(min_litTemp,ALG_WAT_CF[cellLoc])*C_ALG_NUT_CF[cellLoc]; 02002 /* gross production of the 2 communities (gC/m2, NOT kgC/m2) */ 02003 /* density constraint contains both noncalc and calc, competition effect accentuated by calc algae */ 02004 /* used to increase calc growth by factor of 10 */ 02005 NC_ALG_GPP[cellLoc] = NC_ALG_PROD_CF[cellLoc]*GP_ALG_RC_PROD*NC_ALG[cellLoc] 02006 *Max( (1.0-(GP_AlgComp*C_ALG[cellLoc]+NC_ALG[cellLoc])/HP_ALG_MAX[HAB[cellLoc]]),0.0); 02007 C_ALG_GPP[cellLoc] = C_ALG_PROD_CF[cellLoc]*GP_ALG_RC_PROD*C_ALG[cellLoc] 02008 *Max( (1.0-( C_ALG[cellLoc]+NC_ALG[cellLoc])/HP_ALG_MAX[HAB[cellLoc]]),0.0); 02009 /* check for available P mass (the MichMent kinetics nutCF do not) */ 02010 tmp = ( (NC_ALG_GPP[cellLoc]+C_ALG_GPP[cellLoc]) > 0) ? 02011 PO4P / ( (NC_ALG_GPP[cellLoc]+C_ALG_GPP[cellLoc]) 02012 * 0.001*GP_ALG_PC*CELL_SIZE*DT) : 02013 1.0; 02014 if (tmp < 1.0) { 02015 NC_ALG_GPP[cellLoc] *= tmp; 02016 C_ALG_GPP[cellLoc] *= tmp; 02017 /* can have high conc, but low mass of P avail, in presence of high peri biomass and high demand */ 02018 /* reduce the production proportionally if excess demand is found */ 02019 } 02020 /* total of calc and noncalc algae available and their total NPP */ 02021 NC_ALG_NPP[cellLoc] = NC_ALG_GPP[cellLoc]-NC_ALG_RESP[cellLoc]; 02022 C_ALG_NPP[cellLoc] = C_ALG_GPP[cellLoc]-C_ALG_RESP[cellLoc]; 02023 02024 02025 DOM_QUALITY_CF[cellLoc] = (Max(TP_SFWT_CONC_MG[cellLoc],TP_SEDWT_CONCACTMG[cellLoc])) 02026 /GP_DOM_DECOMP_POPT; /* mg/L */ 02027 DOM_TEMP_CF[cellLoc] = Exp(0.20*(H2O_TEMP[cellLoc]-GP_DOM_DECOMP_TOPT))*pow(((40.0-H2O_TEMP[cellLoc])/(40.0-GP_DOM_DECOMP_TOPT)),(0.20*(40.0-GP_DOM_DECOMP_TOPT))); 02028 soil_MOIST_CF[cellLoc] = pow(Max(UNSAT_MOIST_PRP[cellLoc],0.0),0.75); 02029 DOM_DECOMP_POT[cellLoc] = GP_DOM_RCDECOMP*DOM_QUALITY_CF[cellLoc]*DOM_TEMP_CF[cellLoc]*DEPOS_ORG_MAT[cellLoc]*(Min(DOM_SED_AEROB_Z[cellLoc]/HP_DOM_MAXDEPTH[HAB[cellLoc]],1.0)*soil_MOIST_CF[cellLoc]+GP_DOM_DECOMPRED*Min(DOM_SED_ANAEROB_Z[cellLoc]/HP_DOM_MAXDEPTH[HAB[cellLoc]],1.0)); 02030 DOM_DECOMP[cellLoc] = ( (DOM_DECOMP_POT[cellLoc])*DT>DEPOS_ORG_MAT[cellLoc] ) ? ( (DEPOS_ORG_MAT[cellLoc])/DT ) : ( DOM_DECOMP_POT[cellLoc]); 02031 /* added for P DOM stoich */ 02032 DOP_DECOMP[cellLoc] = DOM_DECOMP[cellLoc] * DOM_P_OM[cellLoc]; 02033 02034 SAT_VS_UNSAT[cellLoc] = 1/Exp(100.0*Max((SURFACE_WAT[cellLoc]-UNSAT_DEPTH[cellLoc]),0.0)); 02035 UNSAT_WT_POT[cellLoc] = Max(UNSAT_CAP[cellLoc]-UNSAT_WATER[cellLoc],0.0); 02036 SF_WT_TO_SAT_DOWNFLOW[cellLoc] = ( (1.0-SAT_VS_UNSAT[cellLoc])*UNSAT_WT_POT[cellLoc]*DT>SURFACE_WAT[cellLoc] ) ? ( SURFACE_WAT[cellLoc]/DT ) : ( (1.0-SAT_VS_UNSAT[cellLoc])*UNSAT_WT_POT[cellLoc]); 02037 SAT_WT_RECHG[cellLoc] = ( GP_HYD_RCRECHG*DT>SAT_WATER[cellLoc] ) ? ( SAT_WATER[cellLoc]/DT ) : ( GP_HYD_RCRECHG); 02038 SF_WT_POT_INF[cellLoc] = ( (SURFACE_WAT[cellLoc]<HP_HYD_RCINFILT[HAB[cellLoc]]*DT) ) ? ( SURFACE_WAT[cellLoc]/DT ) : ( HP_HYD_RCINFILT[HAB[cellLoc]]); 02039 SF_WT_INFILTRATION[cellLoc] = ( SF_WT_POT_INF[cellLoc]*SAT_VS_UNSAT[cellLoc]*DT>UNSAT_WT_POT[cellLoc] ) ? ( (UNSAT_WT_POT[cellLoc])/DT ) : ( SF_WT_POT_INF[cellLoc]*SAT_VS_UNSAT[cellLoc]); 02040 HYD_DOM_ACTWAT_PRES[cellLoc] = ( HYD_DOM_ACTWAT_VOL[cellLoc] > 0.01 ) ? ( 1.0 ) : ( 0.0); 02041 HYD_WATER_AVAIL[cellLoc] = Min(1.0, UNSAT_MOIST_PRP[cellLoc]+Exp(-10.0*Max(UNSAT_DEPTH[cellLoc]-HP_NPHBIO_ROOTDEPTH[HAB[cellLoc]],0.0))); 02042 02043 MAC_LIGHT_CF[cellLoc] = SOLRADGRD[cellLoc]/HP_MAC_LIGHTSAT[HAB[cellLoc]]*Exp(1.0-SOLRADGRD[cellLoc]/HP_MAC_LIGHTSAT[HAB[cellLoc]]); 02044 MAC_TEMP_CF[cellLoc] = Exp(0.20*(AIR_TEMP[cellLoc]-HP_MAC_TEMPOPT[HAB[cellLoc]])) *pow(((40.0-AIR_TEMP[cellLoc])/(40.0-HP_MAC_TEMPOPT[HAB[cellLoc]])),(0.20*(40.0-HP_MAC_TEMPOPT[HAB[cellLoc]]))); 02045 MAC_WATER_AVAIL_CF[cellLoc] = graph8(0x0,HYD_WATER_AVAIL[cellLoc]); 02046 MAC_WATER_CF[cellLoc] = Min(MAC_WATER_AVAIL_CF[cellLoc], Max(1.0-Max((SURFACE_WAT[cellLoc]-HP_MAC_WAT_TOLER[HAB[cellLoc]])/HP_MAC_WAT_TOLER[HAB[cellLoc]],0.0),0.0)); 02047 MAC_NUT_CF[cellLoc] = TP_SEDWT_CONCACT[cellLoc]/(TP_SEDWT_CONCACT[cellLoc]+HP_MAC_KSP[HAB[cellLoc]]*0.001) ; 02048 02049 MAC_SALT_CF[cellLoc] = ( HP_MAC_SALIN_THRESH[HAB[cellLoc]]>0.0 ) ? ( Max( 1.0-Max(SAL_SED_WT[cellLoc]-HP_MAC_SALIN_THRESH[HAB[cellLoc]],0.0)/HP_MAC_SALIN_THRESH[HAB[cellLoc]],0.0) ) : ( Max(1.0-SAL_SED_WT[cellLoc],0.0)); 02050 min_litTemp = Min(MAC_LIGHT_CF[cellLoc], MAC_TEMP_CF[cellLoc]); 02051 MAC_PROD_CF[cellLoc] = Min(min_litTemp,MAC_WATER_CF[cellLoc]) 02052 *MAC_NUT_CF[cellLoc]*MAC_SALT_CF[cellLoc]; 02053 PHBIO_NPP[cellLoc] = HP_PHBIO_RCNPP[HAB[cellLoc]]*MAC_PROD_CF[cellLoc]*MAC_PH_BIOMAS[cellLoc]* Max( (1.0-MAC_TOT_BIOM[cellLoc]/MAC_MAX_BIO[cellLoc]), 0.0); 02054 /* check for available P mass that will be taken up (MichMent kinetics in nutCF does not) */ 02055 tmp = (PHBIO_NPP[cellLoc] > 0) ? 02056 TP_SED_WT[cellLoc] / ( PHBIO_NPP[cellLoc] * HP_NPHBIO_IC_PC[HAB[cellLoc]]*CELL_SIZE*DT) : 02057 1.0; 02058 if (tmp < 1.0) PHBIO_NPP[cellLoc] *= tmp; 02059 /* reduce the production proportionally if excess demand is found */ 02060 /* can have high conc, but low mass of P avail, in presence of high plant biomass and high demand */ 02061 /* now add the P and OM associated with that C */ 02062 phbio_npp_P[cellLoc] = PHBIO_NPP[cellLoc] * HP_PHBIO_IC_PC[HAB[cellLoc]]; /* habitat-specfic stoichiometry */ 02063 phbio_npp_OM[cellLoc] = PHBIO_NPP[cellLoc] / HP_PHBIO_IC_CTOOM[HAB[cellLoc]]; /* habitat-specfic stoichiometry */ 02064 02065 /* init ("target") ph/nph ratio and new transloc algorithm */ 02066 MAC_PHtoNPH_Init = HP_PHBIO_MAX[HAB[cellLoc]] / HP_NPHBIO_MAX[HAB[cellLoc]] ; 02067 MAC_PHtoNPH = MAC_PH_BIOMAS[cellLoc] / MAC_NOPH_BIOMAS[cellLoc]; 02068 02069 PHBIO_TRANSLOC_POT[cellLoc] = 0.0; /* (MAC_PHtoNPH<MAC_PHtoNPH_Init) ? (exp(HP_MAC_TRANSLOC_RC[HAB[cellLoc]] *(MAC_PHtoNPH_Init-MAC_PHtoNPH)) - 1.0) : (0.0) */ 02070 02071 PHBIO_TRANSLOC[cellLoc] = ( (PHBIO_TRANSLOC_POT[cellLoc])*DT >NPHBIO_AVAIL[cellLoc] ) ? ( (NPHBIO_AVAIL[cellLoc])/DT ) : ( PHBIO_TRANSLOC_POT[cellLoc]); 02072 /* now remove the P and OM associated with that C */ 02073 phbio_transl_P[cellLoc] = PHBIO_TRANSLOC[cellLoc] * mac_nph_PC[cellLoc]; 02074 phbio_transl_OM[cellLoc] = PHBIO_TRANSLOC[cellLoc] / mac_nph_CtoOM[cellLoc]; 02075 NPHBIO_MORT_POT[cellLoc] = NPHBIO_AVAIL[cellLoc]*HP_PHBIO_RCMORT[HAB[cellLoc]]*(1.0-MAC_PH_BIOMAS[cellLoc]/HP_PHBIO_MAX[HAB[cellLoc]]); 02076 NPHBIO_MORT[cellLoc] = ( (PHBIO_TRANSLOC[cellLoc]+NPHBIO_MORT_POT[cellLoc])*DT>NPHBIO_AVAIL[cellLoc] ) ? ( (NPHBIO_AVAIL[cellLoc]-PHBIO_TRANSLOC[cellLoc]*DT)/DT ) : ( NPHBIO_MORT_POT[cellLoc]); 02077 PHBIO_MORT_POT[cellLoc] = HP_PHBIO_RCMORT[HAB[cellLoc]] *PHBIO_AVAIL[cellLoc] *(1.0-MAC_WATER_AVAIL_CF[cellLoc]); 02078 /* now remove the P and OM associated with that C */ 02079 nphbio_mort_P[cellLoc] = NPHBIO_MORT[cellLoc] * mac_nph_PC[cellLoc]; 02080 nphbio_mort_OM[cellLoc] = NPHBIO_MORT[cellLoc] / mac_nph_CtoOM[cellLoc]; 02081 02082 NPHBIO_TRANSLOC_POT[cellLoc] = 0.0; /* (MAC_PHtoNPH>MAC_PHtoNPH_Init) ? (exp(HP_MAC_TRANSLOC_RC[HAB[cellLoc]] *(MAC_PHtoNPH-MAC_PHtoNPH_Init)) - 1.0) : (0.0) */ 02083 NPHBIO_TRANSLOC[cellLoc] = ( (NPHBIO_TRANSLOC_POT[cellLoc])*DT >MAC_PH_BIOMAS[cellLoc] ) ? ( (MAC_PH_BIOMAS[cellLoc])/DT ) : ( NPHBIO_TRANSLOC_POT[cellLoc]); 02084 /* now remove the P and OM associated with that C */ 02085 nphbio_transl_P[cellLoc] = NPHBIO_TRANSLOC[cellLoc] * mac_ph_PC[cellLoc]; 02086 nphbio_transl_OM[cellLoc] = NPHBIO_TRANSLOC[cellLoc] / mac_ph_CtoOM[cellLoc]; 02087 PHBIO_MORT[cellLoc] = ( (PHBIO_MORT_POT[cellLoc]+NPHBIO_TRANSLOC[cellLoc])*DT>PHBIO_AVAIL[cellLoc] ) ? ( (PHBIO_AVAIL[cellLoc]-NPHBIO_TRANSLOC[cellLoc]*DT)/DT ) : ( PHBIO_MORT_POT[cellLoc]); 02088 /* now remove the P associated with that C */ 02089 phbio_mort_P[cellLoc] = PHBIO_MORT[cellLoc] * mac_ph_PC[cellLoc]; 02090 phbio_mort_OM[cellLoc] = PHBIO_MORT[cellLoc] / mac_ph_CtoOM[cellLoc]; 02091 02092 02093 FLOC_DECOMP_QUAL_CF[cellLoc] = Min(TP_SFWT_CONC_MG[cellLoc]/GP_DOM_DECOMP_POPT,1.0) ; 02094 FLOC_DECOMP_POT[cellLoc] = GP_DOM_RCDECOMP*FLOC[cellLoc]*DOM_TEMP_CF[cellLoc] *FLOC_DECOMP_QUAL_CF[cellLoc]; 02095 FLOC_DECOMP[cellLoc] = ( (FLOC_DECOMP_POT[cellLoc])*DT>FLOC[cellLoc] ) ? ( (FLOC[cellLoc])/DT ) : ( FLOC_DECOMP_POT[cellLoc]); 02096 FLOC_DEPO_POT[cellLoc] = ( SURFACE_WAT[cellLoc] > GP_DetentZ ) ? ( FLOC[cellLoc]*0.05 ) : ( FLOC[cellLoc]/DT); 02097 FLOC_DEPO[cellLoc] = ( (FLOC_DEPO_POT[cellLoc]+FLOC_DECOMP[cellLoc])*DT>FLOC[cellLoc] ) ? ( (FLOC[cellLoc]-FLOC_DECOMP[cellLoc]*DT)/DT ) : ( FLOC_DEPO_POT[cellLoc]); 02098 02099 HYD_MANNINGS_N[cellLoc] = Max(-Abs((HP_MAC_MAXROUGH[HAB[cellLoc]]-HP_MAC_MINROUGH[HAB[cellLoc]])* (pow(2.0,(1.0-SURFACE_WAT[cellLoc]/MAC_HEIGHT[cellLoc]))-1.0)) + HP_MAC_MAXROUGH[HAB[cellLoc]],HP_MAC_MINROUGH[HAB[cellLoc]]); 02100 } /* spatial loop end */ 02101 } /* spatial loop end */ 02102 usrErr("Done."); 02103 02104 } /* end of init_eqns */
| void init_canals | ( | int | irun | ) |
Call to initialize the water managment canal network topology and data.
- Parameters:
-
irun a counter of number of times a new simulation run is executed
Definition at line 2110 of file UnitMod.c.
Referenced by main().
02111 { 02112 if (irun == 1) { 02113 usrErr("Initializing Water Management..."); 02114 Canal_Network_Init(GP_DATUM_DISTANCE,SED_ELEV); /* WatMgmt.c - initialize the canal network topology and data */ 02115 usrErr("Done Water Management."); 02116 } 02117 else { 02118 reinitCanals(); 02119 } 02120 02121 }
| void init_succession | ( | void | ) |
Call to initialize the habitat succession module.
Definition at line 2124 of file UnitMod.c.
Referenced by main().
02125 { 02126 HabSwitch_Init( ); 02127 02128 }
| void PtInterp_read | ( | ) |
On-the-fly interpolations to develop spatial time series from point data.
.
Read the point data into the lists.
Note that this was quickly set up for the ELM Peer Review (2006) project, and (at least in v2.6.0) needs to be used in conjunction with the model experiment parms for synthetic stage boundary conditions (because it skips all gridIO inputs, including SFWMM stage/depth).
For ELM v2.6, extracted/re-used these code fragments from ELM v2.1 code, with associated updated uses/documentation
Definition at line 2680 of file UnitMod.c.
Referenced by main().
02681 { 02682 /* rainfall and pET point data for models that do not use gridIO inputs, but use interpolated point time series */ 02683 /* final arg in PTSL_ReadLists is the column number of of the data (not incl. the 3 date columns) */ 02684 usrErr0 ("\n*** WARNING: You are not using gridIO spatial time series data for rain, pET, and SFWMM-output depth. \nReading rainfall and pET point time series data... "); 02685 pTSList2 = PTSL_Init(50, 2); 02686 PTSL_ReadLists(pTSList2,"rain",TIndex2++,&Timestep2,&NPtTS2, 1); /* rainfall in data-col 1 (file's col 4) */ 02687 02688 pTSList3 = PTSL_Init(50, 2); 02689 PTSL_ReadLists(pTSList3,"pET",TIndex3++,&Timestep3,&NPtTS3, 1); /* pET data in data-col 1 (file's col 4) */ 02690 02691 usrErr("done.\n"); 02692 02693 if (99 == 66) { /* never-get-here: temporary v2.6 PTSL - Below (old code frags extracted from v2.1) are left here as template for other data (or re-implementing the ELM v2.1 pET calculations if data available) */ 02694 pTSList4 = PTSL_Init(50, 2); 02695 PTSL_ReadLists(pTSList4,"pET",TIndex4++,&Timestep4,&NPtTS4, 2); /* dew point temperature data can be in data-col 2 */ 02696 pTSList5 = PTSL_Init(50, 2); 02697 PTSL_ReadLists(pTSList5,"pET",TIndex5++,&Timestep5,&NPtTS5, 3); /* wind speed data can be in data-col 3 */ 02698 pTSList6 = PTSL_Init(50, 2); 02699 PTSL_ReadLists(pTSList6,"pET",TIndex6++,&Timestep6,&NPtTS6, 4); /* cloud cover data can be in data-col 4 */ 02700 } 02701 }
| void reinitBIR | ( | void | ) |
Calls to re-initialize Basin/Indicator-Region data.
Definition at line 2131 of file UnitMod.c.
Referenced by main().
02132 { 02133 usrErr0("Re-initializing Basin/Indicator-Region info..."); 02134 BIRstats_reset(); 02135 BIRbudg_reset(); 02136 Cell_reset_avg(); 02137 Cell_reset_hydper(); 02138 usrErr("Done."); 02139 }
| void reinitCanals | ( | void | ) |
Call to re-initialize canal storages.
Definition at line 2142 of file UnitMod.c.
Referenced by init_canals(), and main().
02143 { 02144 usrErr0("Re-initializing Canal depths & constituent masses..."); 02145 CanalReInit(); 02146 usrErr("Done."); 02147 }
| void alloc_memory | ( | ) |
Allocate memory.
Allocate memory for model variables, and initialize (not context-specific) values.
Definition at line 2717 of file UnitMod.c.
Referenced by main().
02718 { 02719 usrErr0("Allocating Memory..."); /* console message */ 02720 02721 ON_MAP = (unsigned char *) nalloc(sizeof(unsigned char)*(s0+2)*(s1+2),"ON_MAP"); 02722 init_pvar(ON_MAP,NULL,'c',0.0); 02723 02724 BCondFlow = (int *) nalloc(sizeof(int)*(s0+2)*(s1+2),"BCondFlow"); 02725 init_pvar(BCondFlow,NULL,'d',0.0); 02726 HAB = (unsigned char *) nalloc(sizeof(unsigned char)*(s0+2)*(s1+2),"HAB"); 02727 init_pvar(HAB,NULL,'c',0.0); 02728 basn = (int *) nalloc(sizeof(int)*(s0+2)*(s1+2),"basn"); /* Basin/Indicator-Region map */ 02729 init_pvar(basn,NULL,'d',0.0); 02730 02731 ALG_INCID_LIGHT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_INCID_LIGHT"); 02732 init_pvar(ALG_INCID_LIGHT,NULL,'f',0.0); 02733 ALG_LIGHT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_LIGHT_CF"); 02734 init_pvar(ALG_LIGHT_CF,NULL,'f',0.0); 02735 ALG_LIGHT_EXTINCT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_LIGHT_EXTINCT"); 02736 init_pvar(ALG_LIGHT_EXTINCT,NULL,'f',0.0); 02737 ALG_WAT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_WAT_CF"); 02738 init_pvar(ALG_WAT_CF,NULL,'f',0.0); 02739 ALG_TEMP_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_TEMP_CF"); 02740 init_pvar(ALG_TEMP_CF,NULL,'f',0.0); 02741 ALG_REFUGE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_REFUGE"); 02742 init_pvar(ALG_REFUGE,NULL,'f',0.0); 02743 ALG_SAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_SAT"); 02744 init_pvar(ALG_SAT,NULL,'f',0.0); 02745 ALG_TOT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_TOT"); 02746 init_pvar(ALG_TOT,NULL,'f',0.0); 02747 02748 NC_ALG_AVAIL_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_AVAIL_MORT"); 02749 init_pvar(NC_ALG_AVAIL_MORT,NULL,'f',0.0); 02750 NC_ALG_GPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_GPP"); 02751 init_pvar(NC_ALG_GPP,NULL,'f',0.0); 02752 NC_ALG_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_MORT"); 02753 init_pvar(NC_ALG_MORT,NULL,'f',0.0); 02754 NC_ALG_MORT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_MORT_POT"); 02755 init_pvar(NC_ALG_MORT_POT,NULL,'f',0.0); 02756 NC_ALG_NPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_NPP"); 02757 init_pvar(NC_ALG_NPP,NULL,'f',0.0); 02758 NC_ALG_NUT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_NUT_CF"); 02759 init_pvar(NC_ALG_NUT_CF,NULL,'f',0.0); 02760 NC_ALG_PROD_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_PROD_CF"); 02761 init_pvar(NC_ALG_PROD_CF,NULL,'f',0.0); 02762 NC_ALG_RESP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_RESP"); 02763 init_pvar(NC_ALG_RESP,NULL,'f',0.0); 02764 NC_ALG_RESP_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_RESP_POT"); 02765 init_pvar(NC_ALG_RESP_POT,NULL,'f',0.0); 02766 NC_ALG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG"); 02767 init_pvar(NC_ALG,NULL,'f',0.0); 02768 C_ALG_AVAIL_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_AVAIL_MORT"); 02769 init_pvar(C_ALG_AVAIL_MORT,NULL,'f',0.0); 02770 C_ALG_GPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_GPP"); 02771 init_pvar(C_ALG_GPP,NULL,'f',0.0); 02772 C_ALG_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_MORT"); 02773 init_pvar(C_ALG_MORT,NULL,'f',0.0); 02774 C_ALG_MORT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_MORT_POT"); 02775 init_pvar(C_ALG_MORT_POT,NULL,'f',0.0); 02776 C_ALG_NPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_NPP"); 02777 init_pvar(C_ALG_NPP,NULL,'f',0.0); 02778 C_ALG_NUT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_NUT_CF"); 02779 init_pvar(C_ALG_NUT_CF,NULL,'f',0.0); 02780 C_ALG_PROD_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_PROD_CF"); 02781 init_pvar(C_ALG_PROD_CF,NULL,'f',0.0); 02782 C_ALG_RESP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_RESP"); 02783 init_pvar(C_ALG_RESP,NULL,'f',0.0); 02784 C_ALG_RESP_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_RESP_POT"); 02785 init_pvar(C_ALG_RESP_POT,NULL,'f',0.0); 02786 C_ALG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG"); 02787 init_pvar(C_ALG,NULL,'f',0.0); 02788 NC_ALG_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_P"); 02789 init_pvar(NC_ALG_P,NULL,'f',0.0); 02790 C_ALG_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_P"); 02791 init_pvar(C_ALG_P,NULL,'f',0.0); 02792 NC_ALG_GPP_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_GPP_P"); 02793 init_pvar(NC_ALG_GPP_P,NULL,'f',0.0); 02794 C_ALG_GPP_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_GPP_P"); 02795 init_pvar(C_ALG_GPP_P,NULL,'f',0.0); 02796 NC_ALG_MORT_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_MORT_P"); 02797 init_pvar(NC_ALG_MORT_P,NULL,'f',0.0); 02798 C_ALG_MORT_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_MORT_P"); 02799 init_pvar(C_ALG_MORT_P,NULL,'f',0.0); 02800 NC_ALG_PCrep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_PCrep"); 02801 init_pvar(NC_ALG_PCrep,NULL,'f',0.0); 02802 C_ALG_PCrep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_PCrep"); 02803 init_pvar(C_ALG_PCrep,NULL,'f',0.0); 02804 NC_ALG_PC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"NC_ALG_PC"); 02805 init_pvar(NC_ALG_PC,NULL,'l',0.0); 02806 C_ALG_PC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"C_ALG_PC"); 02807 init_pvar(C_ALG_PC,NULL,'l',0.0); 02808 02809 DEPOS_ORG_MAT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"DEPOS_ORG_MAT"); 02810 init_pvar(DEPOS_ORG_MAT,NULL,'l',0.0); 02811 02812 DOM_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_Z"); 02813 init_pvar(DOM_Z,NULL,'f',0.0); 02814 DOM_DECOMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_DECOMP"); 02815 init_pvar(DOM_DECOMP,NULL,'f',0.0); 02816 DOM_DECOMP_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_DECOMP_POT"); 02817 init_pvar(DOM_DECOMP_POT,NULL,'f',0.0); 02818 DOM_fr_nphBio = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_fr_nphBio"); 02819 init_pvar(DOM_fr_nphBio,NULL,'f',0.0); 02820 02821 Floc_fr_phBio = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Floc_fr_phBio"); 02822 init_pvar(Floc_fr_phBio,NULL,'f',0.0); 02823 DOM_FR_FLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_FR_FLOC"); 02824 init_pvar(DOM_FR_FLOC,NULL,'f',0.0); 02825 soil_MOIST_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"soil_MOIST_CF"); 02826 init_pvar(soil_MOIST_CF,NULL,'f',0.0); 02827 DOM_QUALITY_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_QUALITY_CF"); 02828 init_pvar(DOM_QUALITY_CF,NULL,'f',0.0); 02829 DOM_SED_AEROB_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_SED_AEROB_Z"); 02830 init_pvar(DOM_SED_AEROB_Z,NULL,'f',0.0); 02831 DOM_SED_ANAEROB_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_SED_ANAEROB_Z"); 02832 init_pvar(DOM_SED_ANAEROB_Z,NULL,'f',0.0); 02833 DOM_TEMP_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_TEMP_CF"); 02834 init_pvar(DOM_TEMP_CF,NULL,'f',0.0); 02835 ELEVATION = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ELEVATION"); 02836 init_pvar(ELEVATION,NULL,'f',0.0); 02837 Bathymetry = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Bathymetry"); 02838 init_pvar(Bathymetry,NULL,'f',0.0); 02839 SED_ELEV = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SED_ELEV"); 02840 init_pvar(SED_ELEV,NULL,'f',0.0); 02841 SED_INACT_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SED_INACT_Z"); 02842 init_pvar(SED_INACT_Z,NULL,'f',0.0); 02843 DOP_FLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOP_FLOC"); 02844 init_pvar(DOP_FLOC,NULL,'f',0.0); 02845 DOP_nphBio = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOP_nphBio"); 02846 init_pvar(DOP_nphBio,NULL,'f',0.0); 02847 02848 DOM_P_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"DOM_P_OM"); 02849 init_pvar(DOM_P_OM,NULL,'l',0.0); 02850 02851 TPtoSOIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoSOIL"); 02852 init_pvar(TPtoSOIL,NULL,'f',0.0); 02853 TPtoSOIL_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoSOIL_rep"); 02854 init_pvar(TPtoSOIL_rep,NULL,'f',0.0); 02855 TPtoVOL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoVOL"); 02856 init_pvar(TPtoVOL,NULL,'f',0.0); 02857 TPtoVOL_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoVOL_rep"); 02858 init_pvar(TPtoVOL_rep,NULL,'f',0.0); 02859 BulkD = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"BulkD"); 02860 init_pvar(BulkD,NULL,'f',0.0); 02861 DOM_BD = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_BD"); 02862 init_pvar(DOM_BD,NULL,'f',0.0); 02863 Inorg_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Inorg_Z"); 02864 init_pvar(Inorg_Z,NULL,'f',0.0); 02865 DIM = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DIM"); 02866 init_pvar(DIM,NULL,'f',0.0); 02867 DOP_DECOMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOP_DECOMP"); 02868 init_pvar(DOP_DECOMP,NULL,'f',0.0); 02869 02870 DOP = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"DOP"); 02871 init_pvar(DOP,NULL,'l',0.0); 02872 02873 /* placeholder for fire */ 02874 FIREdummy = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FIREdummy"); 02875 init_pvar(FIREdummy,NULL,'f',0.0); 02876 02877 HYD_DOM_ACTWAT_PRES = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_DOM_ACTWAT_PRES"); 02878 init_pvar(HYD_DOM_ACTWAT_PRES,NULL,'f',0.0); 02879 HYD_DOM_ACTWAT_VOL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_DOM_ACTWAT_VOL"); 02880 init_pvar(HYD_DOM_ACTWAT_VOL,NULL,'f',0.0); 02881 HYD_EVAP_CALC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_EVAP_CALC"); 02882 init_pvar(HYD_EVAP_CALC,NULL,'f',0.0); 02883 HYD_MANNINGS_N = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_MANNINGS_N"); 02884 init_pvar(HYD_MANNINGS_N,NULL,'f',0.0); 02885 HYD_RCCONDUCT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_RCCONDUCT"); 02886 init_pvar(HYD_RCCONDUCT,NULL,'f',0.0); 02887 HYD_SAT_POT_TRANS = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_SAT_POT_TRANS"); 02888 init_pvar(HYD_SAT_POT_TRANS,NULL,'f',0.0); 02889 HYD_SED_WAT_VOL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_SED_WAT_VOL"); 02890 init_pvar(HYD_SED_WAT_VOL,NULL,'f',0.0); 02891 HYD_TOT_POT_TRANSP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_TOT_POT_TRANSP"); 02892 init_pvar(HYD_TOT_POT_TRANSP,NULL,'f',0.0); 02893 HydTotHd = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HydTotHd"); 02894 init_pvar(HydTotHd,NULL,'f',0.0); 02895 HydRelDepPosNeg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HydRelDepPosNeg"); 02896 init_pvar(HydRelDepPosNeg,NULL,'f',0.0); 02897 HYD_TRANSP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_TRANSP"); 02898 init_pvar(HYD_TRANSP,NULL,'f',0.0); 02899 HYD_ET = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_ET"); 02900 init_pvar(HYD_ET,NULL,'f',0.0); 02901 HYD_UNSAT_POT_TRANS = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_UNSAT_POT_TRANS"); 02902 init_pvar(HYD_UNSAT_POT_TRANS,NULL,'f',0.0); 02903 HYD_WATER_AVAIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_WATER_AVAIL"); 02904 init_pvar(HYD_WATER_AVAIL,NULL,'f',0.0); 02905 02906 /* sfwmm rainfall, stage, and pET mapped to elm grid */ 02907 boundcond_rain = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"boundcond_rain"); 02908 init_pvar(boundcond_rain,NULL,'f',0.0); 02909 boundcond_depth = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"boundcond_depth"); 02910 init_pvar(boundcond_depth,NULL,'f',0.0); 02911 boundcond_ETp = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"boundcond_ETp"); 02912 init_pvar(boundcond_ETp,NULL,'f',0.0); 02913 02914 SAT_TO_UNSAT_FL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_TO_UNSAT_FL"); 02915 init_pvar(SAT_TO_UNSAT_FL,NULL,'f',0.0); 02916 SAT_VS_UNSAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_VS_UNSAT"); 02917 init_pvar(SAT_VS_UNSAT,NULL,'f',0.0); 02918 SAT_WATER = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_WATER"); 02919 init_pvar(SAT_WATER,NULL,'f',0.0); 02920 SAT_WT_HEAD = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_WT_HEAD"); 02921 init_pvar(SAT_WT_HEAD,NULL,'f',0.0); 02922 SAT_WT_RECHG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_WT_RECHG"); 02923 init_pvar(SAT_WT_RECHG,NULL,'f',0.0); 02924 SAT_WT_TRANSP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_WT_TRANSP"); 02925 init_pvar(SAT_WT_TRANSP,NULL,'f',0.0); 02926 SF_WT_EVAP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_EVAP"); 02927 init_pvar(SF_WT_EVAP,NULL,'f',0.0); 02928 SF_WT_FROM_RAIN = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_FROM_RAIN"); 02929 init_pvar(SF_WT_FROM_RAIN,NULL,'f',0.0); 02930 SF_WT_INFILTRATION = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_INFILTRATION"); 02931 init_pvar(SF_WT_INFILTRATION,NULL,'f',0.0); 02932 SF_WT_POT_INF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_POT_INF"); 02933 init_pvar(SF_WT_POT_INF,NULL,'f',0.0); 02934 SF_WT_TO_SAT_DOWNFLOW = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_TO_SAT_DOWNFLOW"); 02935 init_pvar(SF_WT_TO_SAT_DOWNFLOW,NULL,'f',0.0); 02936 SFWT_VOL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SFWT_VOL"); 02937 init_pvar(SFWT_VOL,NULL,'f',0.0); 02938 SURFACE_WAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SURFACE_WAT"); 02939 init_pvar(SURFACE_WAT,NULL,'f',0.0); 02940 SF_WT_VEL_mag = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_VEL_mag"); 02941 init_pvar(SF_WT_VEL_mag,NULL,'f',0.0); 02942 BCmodel_sfwat = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"BCmodel_sfwat"); 02943 init_pvar(BCmodel_sfwat,NULL,'f',0.0); 02944 02945 UNSAT_AVAIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_AVAIL"); 02946 init_pvar(UNSAT_AVAIL,NULL,'f',0.0); 02947 UNSAT_CAP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_CAP"); 02948 init_pvar(UNSAT_CAP,NULL,'f',0.0); 02949 UNSAT_DEPTH = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_DEPTH"); 02950 init_pvar(UNSAT_DEPTH,NULL,'f',0.0); 02951 UNSAT_HYD_COND_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_HYD_COND_CF"); 02952 init_pvar(UNSAT_HYD_COND_CF,NULL,'f',0.0); 02953 UNSAT_MOIST_PRP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_MOIST_PRP"); 02954 init_pvar(UNSAT_MOIST_PRP,NULL,'f',0.0); 02955 UNSAT_PERC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_PERC"); 02956 init_pvar(UNSAT_PERC,NULL,'f',0.0); 02957 UNSAT_TO_SAT_FL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_TO_SAT_FL"); 02958 init_pvar(UNSAT_TO_SAT_FL,NULL,'f',0.0); 02959 UNSAT_TRANSP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_TRANSP"); 02960 init_pvar(UNSAT_TRANSP,NULL,'f',0.0); 02961 UNSAT_WATER = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_WATER"); 02962 init_pvar(UNSAT_WATER,NULL,'f',0.0); 02963 UNSAT_WT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_WT_POT"); 02964 init_pvar(UNSAT_WT_POT,NULL,'f',0.0); 02965 02966 MAC_HEIGHT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_HEIGHT"); 02967 init_pvar(MAC_HEIGHT,NULL,'f',0.0); 02968 MAC_LAI = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_LAI"); 02969 init_pvar(MAC_LAI,NULL,'f',0.0); 02970 LAI_eff = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"LAI_eff"); 02971 init_pvar(LAI_eff,NULL,'f',0.0); 02972 MAC_LIGHT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_LIGHT_CF"); 02973 init_pvar(MAC_LIGHT_CF,NULL,'f',0.0); 02974 MAC_MAX_BIO = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_MAX_BIO"); 02975 init_pvar(MAC_MAX_BIO,NULL,'f',0.0); 02976 MAC_NOPH_BIOMAS = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_NOPH_BIOMAS"); 02977 init_pvar(MAC_NOPH_BIOMAS,NULL,'f',0.0); 02978 MAC_NUT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_NUT_CF"); 02979 init_pvar(MAC_NUT_CF,NULL,'f',0.0); 02980 MAC_PH_BIOMAS = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_PH_BIOMAS"); 02981 init_pvar(MAC_PH_BIOMAS,NULL,'f',0.0); 02982 MAC_PROD_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_PROD_CF"); 02983 init_pvar(MAC_PROD_CF,NULL,'f',0.0); 02984 MAC_REL_BIOM = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_REL_BIOM"); 02985 init_pvar(MAC_REL_BIOM,NULL,'f',0.0); 02986 MAC_SALT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_SALT_CF"); 02987 init_pvar(MAC_SALT_CF,NULL,'f',0.0); 02988 MAC_TEMP_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_TEMP_CF"); 02989 init_pvar(MAC_TEMP_CF,NULL,'f',0.0); 02990 MAC_TOT_BIOM = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_TOT_BIOM"); 02991 init_pvar(MAC_TOT_BIOM,NULL,'f',0.0); 02992 MAC_WATER_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_WATER_CF"); 02993 init_pvar(MAC_WATER_CF,NULL,'f',0.0); 02994 MAC_WATER_AVAIL_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_WATER_AVAIL_CF"); 02995 init_pvar(MAC_WATER_AVAIL_CF,NULL,'f',0.0); 02996 NPHBIO_AVAIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_AVAIL"); 02997 init_pvar(NPHBIO_AVAIL,NULL,'f',0.0); 02998 NPHBIO_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_MORT"); 02999 init_pvar(NPHBIO_MORT,NULL,'f',0.0); 03000 NPHBIO_MORT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_MORT_POT"); 03001 init_pvar(NPHBIO_MORT_POT,NULL,'f',0.0); 03002 NPHBIO_REFUGE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_REFUGE"); 03003 init_pvar(NPHBIO_REFUGE,NULL,'f',0.0); 03004 NPHBIO_SAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_SAT"); 03005 init_pvar(NPHBIO_SAT,NULL,'f',0.0); 03006 NPHBIO_TRANSLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_TRANSLOC"); 03007 init_pvar(NPHBIO_TRANSLOC,NULL,'f',0.0); 03008 NPHBIO_TRANSLOC_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_TRANSLOC_POT"); 03009 init_pvar(NPHBIO_TRANSLOC_POT,NULL,'f',0.0); 03010 PHBIO_AVAIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_AVAIL"); 03011 init_pvar(PHBIO_AVAIL,NULL,'f',0.0); 03012 PHBIO_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_MORT"); 03013 init_pvar(PHBIO_MORT,NULL,'f',0.0); 03014 PHBIO_MORT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_MORT_POT"); 03015 init_pvar(PHBIO_MORT_POT,NULL,'f',0.0); 03016 PHBIO_NPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_NPP"); 03017 init_pvar(PHBIO_NPP,NULL,'f',0.0); 03018 PHBIO_REFUGE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_REFUGE"); 03019 init_pvar(PHBIO_REFUGE,NULL,'f',0.0); 03020 PHBIO_SAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_SAT"); 03021 init_pvar(PHBIO_SAT,NULL,'f',0.0); 03022 PHBIO_TRANSLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_TRANSLOC"); 03023 init_pvar(PHBIO_TRANSLOC,NULL,'f',0.0); 03024 PHBIO_TRANSLOC_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_TRANSLOC_POT"); 03025 init_pvar(PHBIO_TRANSLOC_POT,NULL,'f',0.0); 03026 03027 phbio_npp_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_npp_P"); 03028 init_pvar(phbio_npp_P,NULL,'l',0.0); 03029 phbio_npp_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_npp_OM"); 03030 init_pvar(phbio_npp_OM,NULL,'l',0.0); 03031 phbio_mort_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_mort_P"); 03032 init_pvar(phbio_mort_P,NULL,'l',0.0); 03033 phbio_mort_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_mort_OM"); 03034 init_pvar(phbio_mort_OM,NULL,'l',0.0); 03035 phbio_transl_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_transl_P"); 03036 init_pvar(phbio_transl_P,NULL,'l',0.0); 03037 phbio_transl_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_transl_OM"); 03038 init_pvar(phbio_transl_OM,NULL,'l',0.0); 03039 nphbio_transl_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"nphbio_transl_P"); 03040 init_pvar(nphbio_transl_P,NULL,'l',0.0); 03041 nphbio_transl_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"nphbio_transl_OM"); 03042 init_pvar(nphbio_transl_OM,NULL,'l',0.0); 03043 nphbio_mort_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"nphbio_mort_P"); 03044 init_pvar(nphbio_mort_P,NULL,'l',0.0); 03045 nphbio_mort_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"nphbio_mort_OM"); 03046 init_pvar(nphbio_mort_OM,NULL,'l',0.0); 03047 mac_nph_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_nph_P"); 03048 init_pvar(mac_nph_P,NULL,'l',0.0); 03049 mac_nph_PC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_nph_PC"); 03050 init_pvar(mac_nph_PC,NULL,'l',0.0); 03051 mac_nph_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_nph_OM"); 03052 init_pvar(mac_nph_OM,NULL,'l',0.0); 03053 mac_nph_CtoOM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_nph_CtoOM"); 03054 init_pvar(mac_nph_CtoOM,NULL,'l',0.0); 03055 mac_ph_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_ph_P"); 03056 init_pvar(mac_ph_P,NULL,'l',0.0); 03057 mac_ph_PC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_ph_PC"); 03058 init_pvar(mac_ph_PC,NULL,'l',0.0); 03059 mac_ph_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_ph_OM"); 03060 init_pvar(mac_ph_OM,NULL,'l',0.0); 03061 mac_ph_CtoOM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_ph_CtoOM"); 03062 init_pvar(mac_ph_CtoOM,NULL,'l',0.0); 03063 03064 mac_nph_PC_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"mac_nph_PC_rep"); 03065 init_pvar(mac_nph_PC_rep,NULL,'f',0.0); 03066 mac_ph_PC_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"mac_ph_PC_rep"); 03067 init_pvar(mac_ph_PC_rep,NULL,'f',0.0); 03068 03069 TP_SED_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SED_WT"); 03070 init_pvar(TP_SED_WT,NULL,'l',0.0); 03071 TP_SED_CONC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SED_CONC"); 03072 init_pvar(TP_SED_CONC,NULL,'l',0.0); 03073 TP_SEDWT_CONCACT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SEDWT_CONCACT"); 03074 init_pvar(TP_SEDWT_CONCACT,NULL,'l',0.0); 03075 TP_SF_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SF_WT"); 03076 init_pvar(TP_SF_WT,NULL,'l',0.0); 03077 TP_SFWT_CONC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SFWT_CONC"); 03078 init_pvar(TP_SFWT_CONC,NULL,'l',0.0); 03079 TP_SORB = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SORB"); 03080 init_pvar(TP_SORB,NULL,'l',0.0); 03081 TP_SORBCONC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SORBCONC"); 03082 init_pvar(TP_SORBCONC,NULL,'l',0.0); 03083 TP_SED_WT_AZ = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SED_WT_AZ"); 03084 init_pvar(TP_SED_WT_AZ,NULL,'l',0.0); 03085 TP_Act_to_Tot = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_Act_to_Tot"); 03086 init_pvar(TP_Act_to_Tot,NULL,'l',0.0); 03087 03088 TP_Act_to_TotRep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_Act_to_TotRep"); 03089 init_pvar(TP_Act_to_TotRep,NULL,'f',0.0); 03090 TP_SORBCONC_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SORBCONC_rep"); 03091 init_pvar(TP_SORBCONC_rep,NULL,'f',0.0); 03092 03093 TP_DNFLOW = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_DNFLOW"); 03094 init_pvar(TP_DNFLOW,NULL,'f',0.0); 03095 TP_DNFLOW_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_DNFLOW_POT"); 03096 init_pvar(TP_DNFLOW_POT,NULL,'f',0.0); 03097 TP_Atm_Depos = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_Atm_Depos"); 03098 init_pvar(TP_Atm_Depos,NULL,'f',0.0); 03099 /* v2.6 added map of atmospheric TP deposition rate */ 03100 TP_AtmosDepos = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_AtmosDepos"); 03101 init_pvar(TP_AtmosDepos,NULL,'f',0.0); 03102 TP_K = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_K"); 03103 init_pvar(TP_K,NULL,'f',0.0); 03104 TP_SED_MINER = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SED_MINER"); 03105 init_pvar(TP_SED_MINER,NULL,'f',0.0); 03106 TP_SEDWT_CONCACTMG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SEDWT_CONCACTMG"); 03107 init_pvar(TP_SEDWT_CONCACTMG,NULL,'f',0.0); 03108 TP_SEDWT_UPTAKE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SEDWT_UPTAKE"); 03109 init_pvar(TP_SEDWT_UPTAKE,NULL,'f',0.0); 03110 TP_SFWT_CONC_MG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SFWT_CONC_MG"); 03111 init_pvar(TP_SFWT_CONC_MG,NULL,'f',0.0); 03112 TP_SFWT_MINER = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SFWT_MINER"); 03113 init_pvar(TP_SFWT_MINER,NULL,'f',0.0); 03114 TP_SFWT_UPTAK = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SFWT_UPTAK"); 03115 init_pvar(TP_SFWT_UPTAK,NULL,'f',0.0); 03116 TP_settl = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_settl"); 03117 init_pvar(TP_settl,NULL,'f',0.0); 03118 TP_SORB_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SORB_POT"); 03119 init_pvar(TP_SORB_POT,NULL,'f',0.0); 03120 TP_SORBTION = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SORBTION"); 03121 init_pvar(TP_SORBTION,NULL,'f',0.0); 03122 TP_UPFLOW = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_UPFLOW"); 03123 init_pvar(TP_UPFLOW,NULL,'f',0.0); 03124 TP_UPFLOW_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_UPFLOW_POT"); 03125 init_pvar(TP_UPFLOW_POT,NULL,'f',0.0); 03126 P_SUM_CELL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"P_SUM_CELL"); 03127 init_pvar(P_SUM_CELL,NULL,'f',0.0); 03128 03129 DINdummy = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"DINdummy"); 03130 init_pvar(DINdummy,NULL,'l',0.0); 03131 03132 WQMsettlVel = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"WQMsettlVel"); 03133 init_pvar(WQMsettlVel,NULL,'f',0.0); 03134 TP_settlDays = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_settlDays"); 03135 init_pvar(TP_settlDays,NULL,'f',0.0); 03136 03137 SAL_SED_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"SAL_SED_WT"); 03138 init_pvar(SAL_SED_WT,NULL,'l',0.0); 03139 SAL_SF_WT_mb = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"SAL_SF_WT_mb"); 03140 init_pvar(SAL_SF_WT_mb,NULL,'l',0.0); 03141 SALT_SED_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"SALT_SED_WT"); 03142 init_pvar(SALT_SED_WT,NULL,'l',0.0); 03143 SALT_SURF_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"SALT_SURF_WT"); 03144 init_pvar(SALT_SURF_WT,NULL,'l',0.0); 03145 03146 SAL_SF_WT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAL_SF_WT"); 03147 init_pvar(SAL_SF_WT,NULL,'f',0.0); 03148 SALT_Atm_Depos = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SALT_Atm_Depos"); 03149 init_pvar(SALT_Atm_Depos,NULL,'f',0.0); 03150 SALT_SED_TO_SF_FLOW = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SALT_SED_TO_SF_FLOW"); 03151 init_pvar(SALT_SED_TO_SF_FLOW,NULL,'f',0.0); 03152 SALT_SFWAT_DOWNFL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SALT_SFWAT_DOWNFL"); 03153 init_pvar(SALT_SFWAT_DOWNFL,NULL,'f',0.0); 03154 SALT_SFWAT_DOWNFL_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SALT_SFWAT_DOWNFL_POT"); 03155 init_pvar(SALT_SFWAT_DOWNFL_POT,NULL,'f',0.0); 03156 03157 FLOC_DECOMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DECOMP"); 03158 init_pvar(FLOC_DECOMP,NULL,'f',0.0); 03159 FLOC_DECOMP_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DECOMP_POT"); 03160 init_pvar(FLOC_DECOMP_POT,NULL,'f',0.0); 03161 FLOC_DECOMP_QUAL_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DECOMP_QUAL_CF"); 03162 init_pvar(FLOC_DECOMP_QUAL_CF,NULL,'f',0.0); 03163 FLOC_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_Z"); 03164 init_pvar(FLOC_Z,NULL,'f',0.0); 03165 FLOC_DEPO = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DEPO"); 03166 init_pvar(FLOC_DEPO,NULL,'f',0.0); 03167 FLOC_DEPO_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DEPO_POT"); 03168 init_pvar(FLOC_DEPO_POT,NULL,'f',0.0); 03169 FLOC_FR_ALGAE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_FR_ALGAE"); 03170 init_pvar(FLOC_FR_ALGAE,NULL,'f',0.0); 03171 FLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC"); 03172 init_pvar(FLOC,NULL,'f',0.0); 03173 FlocP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP"); 03174 init_pvar(FlocP,NULL,'f',0.0); 03175 FlocP_DECOMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_DECOMP"); 03176 init_pvar(FlocP_DECOMP,NULL,'f',0.0); 03177 FlocP_FR_ALGAE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_FR_ALGAE"); 03178 init_pvar(FlocP_FR_ALGAE,NULL,'f',0.0); 03179 FlocP_PhBio = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_PhBio"); 03180 init_pvar(FlocP_PhBio,NULL,'f',0.0); 03181 FlocP_DEPO = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_DEPO"); 03182 init_pvar(FlocP_DEPO,NULL,'f',0.0); 03183 FlocP_OMrep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_OMrep"); 03184 init_pvar(FlocP_OMrep,NULL,'f',0.0); 03185 FlocP_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"FlocP_OM"); 03186 init_pvar(FlocP_OM,NULL,'l',0.0); 03187 03188 SOLRADGRD = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SOLRADGRD"); 03189 init_pvar(SOLRADGRD,NULL,'f',0.0); 03190 SOLRAD274 = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SOLRAD274"); 03191 init_pvar(SOLRAD274,NULL,'f',0.0); 03192 AIR_TEMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"AIR_TEMP"); 03193 init_pvar(AIR_TEMP,NULL,'f',0.0); 03194 H2O_TEMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"H2O_TEMP"); 03195 init_pvar(H2O_TEMP,NULL,'f',0.0); 03196 03197 /* habitat-specific parameter arrays */ 03198 HP_ALG_MAX = NULL; 03199 03200 HP_DOM_MAXDEPTH = NULL; 03201 HP_DOM_AEROBTHIN = NULL; 03202 03203 HP_TP_CONC_GRAD = NULL; 03204 03205 HP_SALT_ICSEDWAT = NULL; 03206 HP_SALT_ICSFWAT = NULL; 03207 03208 HP_PHBIO_MAX = NULL; 03209 HP_NPHBIO_MAX = NULL; 03210 HP_MAC_MAXHT = NULL; 03211 HP_NPHBIO_ROOTDEPTH = NULL; 03212 HP_MAC_MAXROUGH = NULL; 03213 HP_MAC_MINROUGH = NULL; 03214 HP_MAC_MAXLAI = NULL; 03215 HP_MAC_MAXCANOPCOND = NULL; 03216 HP_MAC_CANOPDECOUP = NULL; 03217 HP_MAC_TEMPOPT = NULL; 03218 HP_MAC_LIGHTSAT = NULL; 03219 HP_MAC_KSP = NULL; 03220 HP_PHBIO_RCNPP = NULL; 03221 HP_PHBIO_RCMORT = NULL; 03222 HP_MAC_WAT_TOLER = NULL; 03223 HP_MAC_SALIN_THRESH = NULL; 03224 HP_PHBIO_IC_CTOOM = NULL; 03225 HP_NPHBIO_IC_CTOOM = NULL; 03226 HP_PHBIO_IC_PC = NULL; 03227 HP_NPHBIO_IC_PC = NULL; 03228 HP_MAC_TRANSLOC_RC = NULL; 03229 03230 HP_HYD_RCINFILT = NULL; 03231 HP_HYD_SPEC_YIELD = NULL; 03232 HP_HYD_POROSITY = NULL; 03233 03234 HP_FLOC_IC = NULL; 03235 HP_FLOC_IC_CTOOM = NULL; 03236 HP_FLOC_IC_PC = NULL; 03237 03238 HP_SfDepthLo = NULL; 03239 HP_SfDepthHi = NULL; 03240 HP_SfDepthInt = NULL; 03241 HP_PhosLo = NULL; 03242 HP_PhosHi = NULL; 03243 HP_PhosInt = NULL; 03244 HP_FireInt = NULL; 03245 HP_SalinLo = NULL; 03246 HP_SalinHi = NULL; 03247 HP_SalinInt = NULL; 03248 03249 usrErr("Done."); /* console message */ 03250 03251 }
| void gen_output | ( | int | step, | |
| ViewParm * | view | |||
| ) |
Generate output.
Create output of spatial maps, point locations and debug data (if any)
- Parameters:
-
step The current iteration number view The struct containing output configuration data
- Remarks:
- You need to ensure that the Outlist_size (define in driver_utilities.h) is greater than (at least equal to) the number of output variables here (important if adding variables to this output list).
- Remarks:
- This UnitMod.c code "fragment" (gen_output function, populating tgen[] array) is generated from the "ModelOutlist_creator_v?.xls" OpenOffice workbook. Editing this source directly is not recommended w/o changing ModelOutlist_creator.
The order of the variables in the Model.outList configuration (model input) file MUST EXACTLY match their order in the tgen[] array of the struct.
The ModelOutlist_creator automatically generates both the Model.outList and the tgen[] code.
It is recommended that you utilize the ModelOutlist_creator to generate both the config file and the source code. NOTE: the code generator has a bug, and puts an 'f' argument in all variables, float or not. You must change doubles ('d') and unsigned chars ('c') by hand.
Definition at line 2162 of file UnitMod.c.
Referenced by main().
02163 { 02164 #define numOutputs 50000 02165 static int iw[numOutputs]; 02166 int oIndex; 02167 ViewParm *vp; 02168 02176 static outVar_struct tgen[] = { 02177 { (float**)&TP_settlDays, "TP_settlDays", 'f' }, 02178 { (float**)&FLOC, "FLOC", 'f' }, 02179 { (float**)&FLOC_DECOMP, "FLOC_DECOMP", 'f' }, 02180 { (float**)&FLOC_DECOMP_POT, "FLOC_DECOMP_POT", 'f' }, 02181 { (float**)&FLOC_DECOMP_QUAL_CF, "FLOC_DECOMP_QUAL_CF", 'f' }, 02182 { (float**)&FLOC_DEPO, "FLOC_DEPO", 'f' }, 02183 { (float**)&FLOC_DEPO_POT, "FLOC_DEPO_POT", 'f' }, 02184 { (float**)&FLOC_FR_ALGAE, "FLOC_FR_ALGAE", 'f' }, 02185 { (float**)&FLOC_Z, "FLOC_Z", 'f' }, 02186 { (float**)&FlocP_OMrep, "FlocP_OMrep", 'f' }, 02187 { (float**)&soil_MOIST_CF, "soil_MOIST_CF", 'f' }, 02188 { (float**)&TP_SED_MINER, "TP_SED_MINER", 'f' }, 02189 { (float**)&TP_SFWT_MINER, "TP_SFWT_MINER", 'f' }, 02190 { (float**)&AIR_TEMP, "AIR_TEMP", 'f' }, 02191 { (unsigned char**)&HAB, "HAB", 'c' }, 02192 { (float**)&SOLRAD274, "SOLRAD274", 'f' }, 02193 { (float**)&SOLRADGRD, "SOLRADGRD", 'f' }, 02194 { (float**)&H2O_TEMP, "H2O_TEMP", 'f' }, 02195 { (float**)&HYD_DOM_ACTWAT_PRES, "HYD_DOM_ACTWAT_PRES", 'f' }, 02196 { (float**)&HYD_DOM_ACTWAT_VOL, "HYD_DOM_ACTWAT_VOL", 'f' }, 02197 { (float**)&HYD_ET, "HYD_ET", 'f' }, 02198 { (float**)&HYD_EVAP_CALC, "HYD_EVAP_CALC", 'f' }, 02199 { (float**)&HYD_MANNINGS_N, "HYD_MANNINGS_N", 'f' }, 02200 { (float**)&HYD_SAT_POT_TRANS, "HYD_SAT_POT_TRANS", 'f' }, 02201 { (float**)&HYD_SED_WAT_VOL, "HYD_SED_WAT_VOL", 'f' }, 02202 { (float**)&HYD_TOT_POT_TRANSP, "HYD_TOT_POT_TRANSP", 'f' }, 02203 { (float**)&HYD_TRANSP, "HYD_TRANSP", 'f' }, 02204 { (float**)&HYD_UNSAT_POT_TRANS, "HYD_UNSAT_POT_TRANS", 'f' }, 02205 { (float**)&HYD_WATER_AVAIL, "HYD_WATER_AVAIL", 'f' }, 02206 { (float**)&HydTotHd, "HydTotHd", 'f' }, 02207 { (float**)&HydRelDepPosNeg, "HydRelDepPosNeg", 'f' }, 02208 { (float**)&LAI_eff, "LAI_eff", 'f' }, 02209 { (float**)&MAC_WATER_AVAIL_CF, "MAC_WATER_AVAIL_CF", 'f' }, 02210 { (float**)&SAT_TO_UNSAT_FL, "SAT_TO_UNSAT_FL", 'f' }, 02211 { (float**)&SAT_VS_UNSAT, "SAT_VS_UNSAT", 'f' }, 02212 { (float**)&SAT_WATER, "SAT_WATER", 'f' }, 02213 { (float**)&SAT_WT_HEAD, "SAT_WT_HEAD", 'f' }, 02214 { (float**)&SAT_WT_RECHG, "SAT_WT_RECHG", 'f' }, 02215 { (float**)&SAT_WT_TRANSP, "SAT_WT_TRANSP", 'f' }, 02216 { (float**)&SF_WT_EVAP, "SF_WT_EVAP", 'f' }, 02217 { (float**)&SF_WT_FROM_RAIN, "SF_WT_FROM_RAIN", 'f' }, 02218 { (float**)&SF_WT_INFILTRATION, "SF_WT_INFILTRATION", 'f' }, 02219 { (float**)&SF_WT_POT_INF, "SF_WT_POT_INF", 'f' }, 02220 { (float**)&SF_WT_TO_SAT_DOWNFLOW, "SF_WT_TO_SAT_DOWNFLOW", 'f' }, 02221 { (float**)&SF_WT_VEL_mag, "SF_WT_VEL_mag", 'f' }, 02222 { (float**)&BCmodel_sfwat, "BCmodel_sfwat", 'f' }, 02223 { (float**)&SFWT_VOL, "SFWT_VOL", 'f' }, 02224 { (float**)&SURFACE_WAT, "SURFACE_WAT", 'f' }, 02225 { (float**)&UNSAT_AVAIL, "UNSAT_AVAIL", 'f' }, 02226 { (float**)&UNSAT_CAP, "UNSAT_CAP", 'f' }, 02227 { (float**)&UNSAT_DEPTH, "UNSAT_DEPTH", 'f' }, 02228 { (float**)&UNSAT_HYD_COND_CF, "UNSAT_HYD_COND_CF", 'f' }, 02229 { (float**)&UNSAT_MOIST_PRP, "UNSAT_MOIST_PRP", 'f' }, 02230 { (float**)&UNSAT_PERC, "UNSAT_PERC", 'f' }, 02231 { (float**)&UNSAT_TO_SAT_FL, "UNSAT_TO_SAT_FL", 'f' }, 02232 { (float**)&UNSAT_TRANSP, "UNSAT_TRANSP", 'f' }, 02233 { (float**)&UNSAT_WATER, "UNSAT_WATER", 'f' }, 02234 { (float**)&UNSAT_WT_POT, "UNSAT_WT_POT", 'f' }, 02235 { (float**)&ELEVATION, "ELEVATION", 'f' }, 02236 { (float**)&HYD_RCCONDUCT, "HYD_RCCONDUCT", 'f' }, 02237 { (unsigned char**)&ON_MAP, "ON_MAP", 'c' }, 02238 { (float**)&SED_INACT_Z, "SED_INACT_Z", 'f' }, 02239 { (float**)&MAC_HEIGHT, "MAC_HEIGHT", 'f' }, 02240 { (float**)&MAC_LAI, "MAC_LAI", 'f' }, 02241 { (float**)&MAC_LIGHT_CF, "MAC_LIGHT_CF", 'f' }, 02242 { (float**)&MAC_MAX_BIO, "MAC_MAX_BIO", 'f' }, 02243 { (float**)&MAC_NOPH_BIOMAS, "MAC_NOPH_BIOMAS", 'f' }, 02244 { (float**)&mac_nph_PC_rep, "mac_nph_PC_rep", 'f' }, 02245 { (float**)&MAC_NUT_CF, "MAC_NUT_CF", 'f' }, 02246 { (float**)&MAC_PH_BIOMAS, "MAC_PH_BIOMAS", 'f' }, 02247 { (float**)&mac_ph_PC_rep, "mac_ph_PC_rep", 'f' }, 02248 { (float**)&MAC_PROD_CF, "MAC_PROD_CF", 'f' }, 02249 { (float**)&MAC_REL_BIOM, "MAC_REL_BIOM", 'f' }, 02250 { (float**)&MAC_SALT_CF, "MAC_SALT_CF", 'f' }, 02251 { (float**)&MAC_TEMP_CF, "MAC_TEMP_CF", 'f' }, 02252 { (float**)&MAC_TOT_BIOM, "MAC_TOT_BIOM", 'f' }, 02253 { (float**)&MAC_WATER_CF, "MAC_WATER_CF", 'f' }, 02254 { (float**)&NPHBIO_AVAIL, "NPHBIO_AVAIL", 'f' }, 02255 { (float**)&NPHBIO_MORT, "NPHBIO_MORT", 'f' }, 02256 { (float**)&NPHBIO_MORT_POT, "NPHBIO_MORT_POT", 'f' }, 02257 { (float**)&NPHBIO_REFUGE, "NPHBIO_REFUGE", 'f' }, 02258 { (float**)&NPHBIO_SAT, "NPHBIO_SAT", 'f' }, 02259 { (float**)&NPHBIO_TRANSLOC, "NPHBIO_TRANSLOC", 'f' }, 02260 { (float**)&NPHBIO_TRANSLOC_POT, "NPHBIO_TRANSLOC_POT", 'f' }, 02261 { (float**)&PHBIO_AVAIL, "PHBIO_AVAIL", 'f' }, 02262 { (float**)&PHBIO_MORT, "PHBIO_MORT", 'f' }, 02263 { (float**)&PHBIO_MORT_POT, "PHBIO_MORT_POT", 'f' }, 02264 { (float**)&PHBIO_NPP, "PHBIO_NPP", 'f' }, 02265 { (float**)&PHBIO_REFUGE, "PHBIO_REFUGE", 'f' }, 02266 { (float**)&PHBIO_SAT, "PHBIO_SAT", 'f' }, 02267 { (float**)&PHBIO_TRANSLOC, "PHBIO_TRANSLOC", 'f' }, 02268 { (float**)&PHBIO_TRANSLOC_POT, "PHBIO_TRANSLOC_POT", 'f' }, 02269 { (float**)&TP_SEDWT_UPTAKE, "TP_SEDWT_UPTAKE", 'f' }, 02270 { (float**)&ALG_INCID_LIGHT, "ALG_INCID_LIGHT", 'f' }, 02271 { (float**)&ALG_LIGHT_CF, "ALG_LIGHT_CF", 'f' }, 02272 { (float**)&ALG_LIGHT_EXTINCT, "ALG_LIGHT_EXTINCT", 'f' }, 02273 { (float**)&ALG_REFUGE, "ALG_REFUGE", 'f' }, 02274 { (float**)&ALG_SAT, "ALG_SAT", 'f' }, 02275 { (float**)&ALG_TEMP_CF, "ALG_TEMP_CF", 'f' }, 02276 { (float**)&ALG_TOT, "ALG_TOT", 'f' }, 02277 { (float**)&ALG_WAT_CF, "ALG_WAT_CF", 'f' }, 02278 { (float**)&C_ALG, "C_ALG", 'f' }, 02279 { (float**)&C_ALG_AVAIL_MORT, "C_ALG_AVAIL_MORT", 'f' }, 02280 { (float**)&C_ALG_GPP, "C_ALG_GPP", 'f' }, 02281 { (float**)&C_ALG_MORT, "C_ALG_MORT", 'f' }, 02282 { (float**)&C_ALG_MORT_POT, "C_ALG_MORT_POT", 'f' }, 02283 { (float**)&C_ALG_NPP, "C_ALG_NPP", 'f' }, 02284 { (float**)&C_ALG_NUT_CF, "C_ALG_NUT_CF", 'f' }, 02285 { (float**)&C_ALG_PROD_CF, "C_ALG_PROD_CF", 'f' }, 02286 { (float**)&C_ALG_RESP, "C_ALG_RESP", 'f' }, 02287 { (float**)&C_ALG_RESP_POT, "C_ALG_RESP_POT", 'f' }, 02288 { (float**)&NC_ALG, "NC_ALG", 'f' }, 02289 { (float**)&NC_ALG_AVAIL_MORT, "NC_ALG_AVAIL_MORT", 'f' }, 02290 { (float**)&NC_ALG_GPP, "NC_ALG_GPP", 'f' }, 02291 { (float**)&NC_ALG_MORT, "NC_ALG_MORT", 'f' }, 02292 { (float**)&NC_ALG_MORT_POT, "NC_ALG_MORT_POT", 'f' }, 02293 { (float**)&NC_ALG_NPP, "NC_ALG_NPP", 'f' }, 02294 { (float**)&NC_ALG_NUT_CF, "NC_ALG_NUT_CF", 'f' }, 02295 { (float**)&NC_ALG_PROD_CF, "NC_ALG_PROD_CF", 'f' }, 02296 { (float**)&NC_ALG_RESP, "NC_ALG_RESP", 'f' }, 02297 { (float**)&NC_ALG_RESP_POT, "NC_ALG_RESP_POT", 'f' }, 02298 { (float**)&TP_SFWT_UPTAK, "TP_SFWT_UPTAK", 'f' }, 02299 { (float**)&TP_Act_to_TotRep, "TP_Act_to_TotRep", 'f' }, 02300 { (float**)&TP_DNFLOW, "TP_DNFLOW", 'f' }, 02301 { (float**)&TP_DNFLOW_POT, "TP_DNFLOW_POT", 'f' }, 02302 { (float**)&TP_Atm_Depos, "TP_Atm_Depos", 'f' }, 02303 { (float**)&TP_K, "TP_K", 'f' }, 02304 { (double**)&TP_SED_CONC, "TP_SED_CONC", 'd' }, 02305 { (double**)&TP_SED_WT, "TP_SED_WT", 'd' }, 02306 { (double**)&TP_SED_WT_AZ, "TP_SED_WT_AZ", 'd' }, 02307 { (double**)&TP_SEDWT_CONCACT, "TP_SEDWT_CONCACT", 'd' }, 02308 { (float**)&TP_SEDWT_CONCACTMG, "TP_SEDWT_CONCACTMG", 'f' }, 02309 { (float**)&TP_settl, "TP_settl", 'f' }, 02310 { (double**)&TP_SF_WT, "TP_SF_WT", 'd' }, 02311 { (double**)&TP_SFWT_CONC, "TP_SFWT_CONC", 'd' }, 02312 { (float**)&TP_SFWT_CONC_MG, "TP_SFWT_CONC_MG", 'f' }, 02313 { (double**)&TP_SORB, "TP_SORB", 'd' }, 02314 { (float**)&TP_SORB_POT, "TP_SORB_POT", 'f' }, 02315 { (double**)&TP_SORBCONC, "TP_SORBCONC", 'd' }, 02316 { (float**)&TP_SORBCONC_rep, "TP_SORBCONC_rep", 'f' }, 02317 { (float**)&TP_SORBTION, "TP_SORBTION", 'f' }, 02318 { (float**)&TP_UPFLOW, "TP_UPFLOW", 'f' }, 02319 { (float**)&TP_UPFLOW_POT, "TP_UPFLOW_POT", 'f' }, 02320 { (double**)&SAL_SED_WT, "SAL_SED_WT", 'd' }, 02321 { (float**)&SAL_SF_WT, "SAL_SF_WT", 'f' }, 02322 { (float**)&SALT_SED_TO_SF_FLOW, "SALT_SED_TO_SF_FLOW", 'f' }, 02323 { (double**)&SALT_SED_WT, "SALT_SED_WT", 'd' }, 02324 { (float**)&SALT_SFWAT_DOWNFL, "SALT_SFWAT_DOWNFL", 'f' }, 02325 { (float**)&SALT_SFWAT_DOWNFL_POT, "SALT_SFWAT_DOWNFL_POT", 'f' }, 02326 { (double**)&SALT_SURF_WT, "SALT_SURF_WT", 'd' }, 02327 { (float**)&SALT_Atm_Depos, "SALT_Atm_Depos", 'f' }, 02328 { (double**)&DEPOS_ORG_MAT, "DEPOS_ORG_MAT", 'd' }, 02329 { (float**)&DOM_DECOMP, "DOM_DECOMP", 'f' }, 02330 { (float**)&DOM_DECOMP_POT, "DOM_DECOMP_POT", 'f' }, 02331 { (float**)&DOM_FR_FLOC, "DOM_FR_FLOC", 'f' }, 02332 { (double**)&DOM_P_OM, "DOM_P_OM", 'd' }, 02333 { (float**)&DOM_QUALITY_CF, "DOM_QUALITY_CF", 'f' }, 02334 { (float**)&DOM_SED_AEROB_Z, "DOM_SED_AEROB_Z", 'f' }, 02335 { (float**)&DOM_SED_ANAEROB_Z, "DOM_SED_ANAEROB_Z", 'f' }, 02336 { (float**)&DOM_TEMP_CF, "DOM_TEMP_CF", 'f' }, 02337 { (float**)&DOM_Z, "DOM_Z", 'f' }, 02338 { (double**)&DOP, "DOP", 'd' }, 02339 { (float**)&DOP_DECOMP, "DOP_DECOMP", 'f' }, 02340 { (float**)&P_SUM_CELL, "P_SUM_CELL", 'f' }, 02341 { (float**)&SED_ELEV, "SED_ELEV", 'f' }, 02342 { (float**)&TPtoSOIL_rep, "TPtoSOIL_rep", 'f' }, 02343 { (float**)&Floc_fr_phBioAvg, "Floc_fr_phBioAvg", 'f' }, 02344 { (float**)&TPSfMinAvg, "TPSfMinAvg", 'f' }, 02345 { (float**)&ETAvg, "ETAvg", 'f' }, 02346 { (float**)&EvapAvg, "EvapAvg", 'f' }, 02347 { (float**)&HydPerAnn, "HydPerAnn", 'f' }, 02348 { (float**)&LAI_effAvg, "LAI_effAvg", 'f' }, 02349 { (float**)&Manning_nAvg, "Manning_nAvg", 'f' }, 02350 { (float**)&RainAvg, "RainAvg", 'f' }, 02351 { (float**)&SfWatAvg, "SfWatAvg", 'f' }, 02352 { (float**)&TotHeadAvg, "TotHeadAvg", 'f' }, 02353 { (float**)&HydRelDepPosNegAvg, "HydRelDepPosNegAvg", 'f' }, 02354 { (float**)&TranspAvg, "TranspAvg", 'f' }, 02355 { (float**)&UnsatMoistAvg, "UnsatMoistAvg", 'f' }, 02356 { (float**)&UnsatZavg, "UnsatZavg", 'f' }, 02357 { (float**)&SF_WT_VEL_magAvg, "SF_WT_VEL_magAvg", 'f' }, 02358 { (float**)&BCmodel_sfwatAvg, "BCmodel_sfwatAvg", 'f' }, 02359 { (float**)&mac_nph_PCAvg, "mac_nph_PCAvg", 'f' }, 02360 { (float**)&Mac_nphBioAvg, "Mac_nphBioAvg", 'f' }, 02361 { (float**)&Mac_nphMortAvg, "Mac_nphMortAvg", 'f' }, 02362 { (float**)&Mac_nppAvg, "Mac_nppAvg", 'f' }, 02363 { (float**)&mac_ph_PCAvg, "mac_ph_PCAvg", 'f' }, 02364 { (float**)&Mac_phBioAvg, "Mac_phBioAvg", 'f' }, 02365 { (float**)&Mac_phMortAvg, "Mac_phMortAvg", 'f' }, 02366 { (float**)&Mac_totBioAvg, "Mac_totBioAvg", 'f' }, 02367 { (float**)&MacNutCfAvg, "MacNutCfAvg", 'f' }, 02368 { (float**)&MacWatCfAvg, "MacWatCfAvg", 'f' }, 02369 { (float**)&TPSedUptAvg, "TPSedUptAvg", 'f' }, 02370 { (float**)&C_Peri_mortAvg, "C_Peri_mortAvg", 'f' }, 02371 { (float**)&C_Peri_nppAvg, "C_Peri_nppAvg", 'f' }, 02372 { (float**)&C_Peri_PCAvg, "C_Peri_PCAvg", 'f' }, 02373 { (float**)&C_PeriAvg, "C_PeriAvg", 'f' }, 02374 { (float**)&C_PeriNutCFAvg, "C_PeriNutCFAvg", 'f' }, 02375 { (float**)&C_PeriRespAvg, "C_PeriRespAvg", 'f' }, 02376 { (float**)&NC_Peri_mortAvg, "NC_Peri_mortAvg", 'f' }, 02377 { (float**)&NC_Peri_nppAvg, "NC_Peri_nppAvg", 'f' }, 02378 { (float**)&NC_Peri_PCAvg, "NC_Peri_PCAvg", 'f' }, 02379 { (float**)&NC_PeriAvg, "NC_PeriAvg", 'f' }, 02380 { (float**)&NC_PeriNutCFAvg, "NC_PeriNutCFAvg", 'f' }, 02381 { (float**)&NC_PeriRespAvg, "NC_PeriRespAvg", 'f' }, 02382 { (float**)&PeriAvg, "PeriAvg", 'f' }, 02383 { (float**)&PeriLiteCFAvg, "PeriLiteCFAvg", 'f' }, 02384 { (float**)&TPSfUptAvg, "TPSfUptAvg", 'f' }, 02385 { (float**)&TP_settlAvg, "TP_settlAvg", 'f' }, 02386 { (float**)&TPSedWatAvg, "TPSedWatAvg", 'f' }, 02387 { (float**)&TPSfWatAvg, "TPSfWatAvg", 'f' }, 02388 { (float**)&SaltSedAvg, "SaltSedAvg", 'f' }, 02389 { (float**)&SaltSfAvg, "SaltSfAvg", 'f' }, 02390 { (float**)&SedElevAvg, "SedElevAvg", 'f' }, 02391 { (float**)&TPSedMinAvg, "TPSedMinAvg", 'f' }, 02392 { (float**)&TPSorbAvg, "TPSorbAvg", 'f' }, 02393 { (float**)&TPtoSOILAvg, "TPtoSOILAvg", 'f' }, 02394 { (float**)&TPtoVOLAvg, "TPtoVOLAvg", 'f' }, 02395 02396 02397 02398 02399 02400 02401 02402 02403 { NULL, NULL, '\0' }, 02404 }; 02405 02406 outVar_struct *ptable; 02407 int i; 02408 02409 for (i = 0, ptable = tgen; ptable->pfvar != NULL; ptable++, i++) 02410 { 02411 vp = view + i; 02412 02413 /* TODO: develop flexible output of calendar-based and julian-based outsteps (jan 11, 2005) */ 02414 if (vp->step > 0) 02415 02416 if (strcmp(ptable->pname,"HydPerAnn")!=0) { /* i.e., not the HydPerAnn variable */ 02417 02418 if (step % vp->step == 0 && (vp->step != CalMonOut) ) { /* standard julian-day outstep interval variables (note: the != CalMonOut needed for step=0) */ 02419 oIndex = iw[i]++; 02420 write_output(oIndex, vp, *(ptable->pfvar), ptable->pname, ptable->ctype, step); 02421 } 02422 else if ( (avgPrint) && (vp->step == CalMonOut) ) { /* variables output at the special 1-calendar-month outstep interval */ 02423 oIndex = iw[i]++; 02424 write_output(oIndex, vp, *(ptable->pfvar), ptable->pname, ptable->ctype, step); 02425 } 02426 } 02427 02428 else 02429 if (FMOD(DAYJUL, 273.0) ==0) { /* hydroperiod is printed at a special-case time (approximately Oct 1 every year) */ 02430 oIndex = iw[i]++; 02431 write_output(oIndex, vp, *(ptable->pfvar), ptable->pname, ptable->ctype, step); 02432 } 02433 02434 02435 } 02436 02437 /* after printing, zero the arrays holding averages or hydroperiods (v2.2.1note using avgPrint var)*/ 02438 if (avgPrint) { 02439 Cell_reset_avg(); 02440 } 02441 02442 if (FMOD(DAYJUL, 273.0) ==0) { 02443 Cell_reset_hydper(); 02444 } 02445 02446 02447 } /* end of gen_output routine */
| void get_map_dims | ( | ) |
Get the map dimensions of the global model array.
This mererly calls the "read_map_dims" function
Definition at line 2707 of file UnitMod.c.
Referenced by setup().
02708 { 02709 read_map_dims("Elevation"); 02710 }
| int call_cell_dyn | ( | int | sector, | |
| int | step | |||
| ) |
Calling function for the cell_dyn** dynamic ecological modules.
This function calls the cell_dyn dynamic modules as defined in the Driver.parm data file. Normal order for calling ELM modules(Sectors): 1 0 7 10 9 2 8 4 12 99.
S#0 hydro: cell-cell horiz (&canals if WaterManagement is on)
S#1 global forcings
S#2 algae/periphyton
S#4 DOM/DOP
S#7 hydro: vertical
S#8 macrophytes
S#9 phosphorus
S#10 salt
S#12 FLOC
S#13 TP net settling
S#99 mass balance, budget, avg, hydroperiod, etc calculations
- Returns:
- rv (=1)
- Parameters:
-
sector The number of the cell_dyn** module being called step The current iteration number
Definition at line 150 of file UnitMod.c.
Referenced by main().
00151 { 00152 int rv=0; 00153 00154 switch(sector) { 00155 00156 case 99: { stats(step); rv=1; } break; 00157 case 0: { horizFlow(step); rv=1; } break; 00158 case 1: { cell_dyn1(step); rv=1; } break; 00159 case 2: { cell_dyn2(step); rv=1; } break; 00160 case 4: { cell_dyn4(step); rv=1; } break; 00161 case 7: { cell_dyn7(step); rv=1; } break; 00162 case 8: { cell_dyn8(step); rv=1; } break; 00163 case 9: { cell_dyn9(step); rv=1; } break; 00164 case 10: { cell_dyn10(step); rv=1; } break; 00165 case 12: { cell_dyn12(step); rv=1; } break; 00166 case 13: { cell_dyn13(step); rv=1; } break; 00167 default: printf("Warning, undefined sector number:%d\n",sector); 00168 } 00169 return rv; 00170 }
Variable Documentation
| int NSector |
number of Sectors (dynamic cell_dyn modules)
Definition at line 28 of file generic_driver.h.
Referenced by get_parmf(), and main().
| int iSector[MAX_SECTOR] |
sector (dynamic cell_dyn module) number
Definition at line 29 of file generic_driver.h.
Referenced by get_parmf(), and main().
| int seed |
the seed value for (pseudo-) random number generator (unused)
Definition at line 30 of file generic_driver.h.
Referenced by get_parmf(), and local_setup().
| int ESPmodeON = 0 |
boolean flag indicating Everglades Settling-of Phosphorus mode. A mode with all biol/chem (non-hydro) modules turned off, running only a net settling rate module that reproduces equations and data from the SFWMD's old Everglades Water Quality Model (EWQM). Done only for comparision to full ecological ELM - ESP doesn't work very well!!
Definition at line 31 of file generic_driver.h.
Referenced by BIRbudg_date(), BIRbudg_print(), BIRbudg_reset(), BIRbudg_sum(), BIRbudg_sumFinal(), BIRoutfiles(), get_parmf(), init_static_data(), and main().
| int WatMgmtOn |
boolean flag indicating water management mode
Definition at line 34 of file generic_driver.h.
Referenced by Flux_SWstuff(), get_parmf(), horizFlow(), and main().
| int HabSwitchOn |
boolean flag indicating habitat succession mode
Definition at line 35 of file generic_driver.h.
Referenced by cell_dyn1(), get_parmf(), and main().
| int PositAnalOn = 0 |
flag for Position Analysis mode: for probability analysis of near-term water management options, Position Analysis mode involves re-inititialization of model on particular month/day at recurring yearly interval
Definition at line 36 of file generic_driver.h.
Referenced by get_parmf(), and main().
| int IsSubstituteModel = 0 |
flag to indicate whether user is asking to substitute the Boundary Condition Model's "stage_minus_landElevation" variable into ELM hydro variables, running only the cell_dyn1 vertical solution module of ELM. This is used to post-process the SFWMM or NSM using the same procedures as ELM
Definition at line 38 of file generic_driver.h.
Referenced by cell_dyn1(), and main().
| char SimAlt[30] |
simulation scenario/alterative name
Definition at line 42 of file generic_driver.h.
Referenced by BIRoutfiles(), Canal_Network_Init(), get_parmf(), open_point_lists(), Read_schedule(), ReadChanStruct(), ReadStructFlow_PtSer(), ReadStructTP_PtSer(), ReadStructTS_PtSer(), ReadStructures(), and writeFloatNetCDF().
| char SimModif[30] |
simulation scenario/alterative modifier name/note
Definition at line 43 of file generic_driver.h.
Referenced by BIRoutfiles(), Canal_Network_Init(), get_parmf(), open_point_lists(), ReadStructures(), and writeFloatNetCDF().
| char outpath[120] |
base pathname for all output
Definition at line 44 of file generic_driver.h.
Referenced by get_parmf().
| int gbl_size[2] |
row [0] and column [1] dimensions of model grid domain, serial implementation
Definition at line 46 of file generic_driver.h.
Referenced by read_header(), read_map_dims(), readMap(), setup_grid(), write_header(), writeFloatMap(), and writeMap().
| char initDateRead[15] |
calendar date of simulation initialization
Definition at line 48 of file generic_driver.h.
Referenced by formatDate(), get_parmf(), and writeFloatNetCDF().
| double Jdate_init |
julian day (since epochs ago) of the initialization of model
Definition at line 49 of file generic_driver.h.
Referenced by get_parmf(), readSeriesCol(), ReadStructFlow_PtSer(), ReadStructTP_PtSer(), ReadStructTS_PtSer(), send_point_lists2(), and track_time().
| double Jdate_end |
julian day (since epochs ago) of the ending of model
Definition at line 50 of file generic_driver.h.
Referenced by get_parmf(), ReadStructFlow_PtSer(), ReadStructTP_PtSer(), and ReadStructTS_PtSer().
| int yr_in |
- Variables associated with the gregorian calendar
yr_in calendar year of initialization of model
mo_in calendar month of initialization of model
da_in calendar day of initialization of model
yr_end calendar year of ending of model
mo_end calendar month of ending of model
da_end calendar day of ending of model
hr_in hour of initialization of model (unused)
mi_in min of initialization of model (unused)
se_in seconds of initialization of model (unused)
Definition at line 62 of file generic_driver.h.
Referenced by get_parmf(), and main().
| int mo_in |
| int da_in |
| int hr_in |
| int mi_in |
| int se_in |
| int yr_end |
| int mo_end |
| int da_end |
| int mo_R_in |
month of Re-initialization in Position analysis mode
Definition at line 63 of file generic_driver.h.
Referenced by get_parmf(), and main().
| int da_R_in |
day of Re-initialization in Position analysis mode
Definition at line 64 of file generic_driver.h.
Referenced by get_parmf(), and main().
| int PORnumday |
number of days of simulation Period Of Record (incl. start day)
Definition at line 65 of file generic_driver.h.
Referenced by get_parmf(), processData(), readSeriesCol(), ReadStructFlow_PtSer(), ReadStructTP_PtSer(), ReadStructTS_PtSer(), and track_time().
| int N_iter |
count of maximum number of model time iterations
Definition at line 67 of file generic_driver.h.
Referenced by get_parmf(), main(), print_point(), and track_time().
| int istep |
counter for number of model time iterations
Definition at line 68 of file generic_driver.h.
Referenced by main(), and quick_look().
| float step_Cell |
constant used in horizontal surface water raster flux equations ( m^(-1.5) * sec )
Definition at line 69 of file generic_driver.h.
Referenced by Flux_SWcells(), and get_parmf().
| float avg_Intvl = 0 |
time (day) interval between recurring-average summaries
Definition at line 71 of file generic_driver.h.
Referenced by CellAvg(), get_parmf(), readViewParms(), and track_time().
| float budg_Intvl = 0 |
time (day) interval between budget summaries
Definition at line 72 of file generic_driver.h.
Referenced by BIRbudg_print(), get_parmf(), and track_time().
| float BIRavg_Intvl = 0 |
time (day) interval between Basin/Indicator-Region stat summaries
Definition at line 73 of file generic_driver.h.
Referenced by BIRstats_print(), and track_time().
| float BIRhyd_avg_Intvl = 0 |
time (day) interval between Basin/Indicator-Region hydro stat summaries
Definition at line 74 of file generic_driver.h.
Referenced by BIRstats_hydro_print(), get_parmf(), and track_time().
| float can_Intvl = 0 |
time (day) interval between canal (water management) summaries
Definition at line 75 of file generic_driver.h.
Referenced by get_parmf(), and track_time().
| int budgCalendar |
boolean flag signifying use of gregorian calendar for budget and BIRavg calcs
Definition at line 77 of file generic_driver.h.
Referenced by get_parmf(), and track_time().
| int BIRhydCalendar |
boolean flag signifying use of gregorian calendar for BIR hydro avg calcs
Definition at line 78 of file generic_driver.h.
Referenced by get_parmf(), and track_time().
| int avgCalendar = 0 |
boolean flag signifying use of gregorian calendar for recurring-average calcs
Definition at line 79 of file generic_driver.h.
Referenced by get_parmf(), and track_time().
| int canalCalendar = 0 |
boolean flag signifying use of gregorian calendar for canal data output
Definition at line 80 of file generic_driver.h.
Referenced by get_parmf(), and track_time().
| int avgPrint = 0 |
boolean flag to indicate if recurring-averages is to be printed at current time
Definition at line 81 of file generic_driver.h.
Referenced by CellAvg(), gen_output(), and track_time().
| int canPrint = 0 |
boolean flag to indicate if canal data is to be printed at current time
Definition at line 82 of file generic_driver.h.
Referenced by Run_Canal_Network(), and track_time().
Definition at line 86 of file generic_driver.h.
Referenced by alloc_mem_runs(), BIRstats_date(), get_global_parm(), get_hab_parm(), get_modexperim_parm(), get_parmf(), main(), and SensiParm_list().
Definition at line 87 of file generic_driver.h.
Referenced by alloc_mem_runs(), get_parmf(), main(), and SensiParm_list().
| float gRTable[] |
Initial value:
{.5000,.5398,.5793,.6179,.6554,.6915,.7257,.7580,.7881,.8159,
.8413,.8643,.8849,.9032,.9192,.9332,.9452,.9554,.9641,.9713,
.9773,.9821,.9861,.9893,.9918,.9938,.9953,.9965,.9974,.9981,
.9987,.9990,.9993,.9995,.9997,.9998,.9998,.9999,.9999,1.00}
Definition at line 89 of file generic_driver.h.
Referenced by Normal().
| SeriesParm pSeries[MAX_PTSERIES] |
An array of SeriesParm structs for point time series output
Definition at line 96 of file generic_driver.h.
Referenced by get_parmf(), main(), print_point(), and setup().
struct of type Point2D, the grid location of a point
Definition at line 97 of file generic_driver.h.
Referenced by get_parmf(), read_map_file(), and write_map_file().
struct of nodenv, with parallel code info
Definition at line 99 of file generic_driver.h.
Referenced by setup_platform().
| int procnum |
- Data for parallel implementation, basically ignore
Lprocnum NA for serial implementation (=1)
procnum NA for serial implementation (also see nodenv struct)
nprocs NA for serial implementation (also see nodenv struct)
recpnum NA for serial implementation
tramType NA for serial implementation
tramNum NA for serial implementation
lcl_size used in serial code, but not really pertinent: in serial implementation, rows lcl_size[0]=s0 and columns lcl_size[1]=s1, dimensions of model grid domain local to one processor (for parallel implementation)
lcl_start used in serial code, but not really pertinent: in serial implementation, starting row lcl_start[0]=0 and starting column lcl_start[1]=0 in model grid domain local to one processor (for parallel implementation)
Definition at line 111 of file generic_driver.h.
Referenced by get_parmf(), local_setup(), main(), read_map_file(), setup_grid(), and setup_platform().
| int Lprocnum |
Definition at line 111 of file generic_driver.h.
Referenced by local_setup(), PTSL_ReadLists(), and read_map_dims().
| int nprocs[2] |
Definition at line 111 of file generic_driver.h.
Referenced by exgridsize(), local_setup(), and setup_platform().
| int recpnum[2] |
Definition at line 111 of file generic_driver.h.
Referenced by exgridsize(), local_setup(), quick_look(), and setup_platform().
| int tramType |
| int tramNum[2] |
| int lcl_size[2] |
| int lcl_start[2] |
Definition at line 111 of file generic_driver.h.
Referenced by print_loc_ave(), print_point(), PTSL_GetInterpolatedValue0(), quick_look(), and setup_grid().
| char* ModelPath |
- Environment variables used in model
ModelPath environment variable, base pathname for executable and input data
ProjName environment variable, the name of the model project
DriverPath environment variable, base pathname for source code
OS_TYPE environment variable, the type of operating system being used (informational purpose only, not used in code)
Definition at line 40 of file driver_utilities.h.
| char * ProjName |
Definition at line 40 of file driver_utilities.h.
| char * OS_TYPE |
| char* OutputPath |
base pathname for all model output (user input)
Definition at line 42 of file driver_utilities.h.
Referenced by BIRoutfiles(), Canal_Network_Init(), cell_dyn1(), Channel_configure(), get_parmf(), main(), open_debug_outFile(), open_point_lists(), send_point_lists2(), write_map_file(), and writeSeries().
| char modelName[20] |
- Model name/version (user input)
modelName Name given to model implementation (user input)
modelVers Version given to model implementation (e.g., v.2.1) (user input)
Definition at line 48 of file driver_utilities.h.
Referenced by BIRinit(), BIRoutfiles(), Canal_Network_Init(), get_parmf(), open_point_lists(), Read_schedule(), ReadChanStruct(), ReadStructures(), and writeFloatNetCDF().
| char modelVers[10] |
Definition at line 48 of file driver_utilities.h.
Referenced by BIRoutfiles(), Canal_Network_Init(), get_parmf(), open_point_lists(), and writeFloatNetCDF().
 1.5.6
1.5.6