unitmod.h File Reference
Header file for the ecological Unit Model. More...
#include "globals.h"
Go to the source code of this file.
Functions | |
| int | call_cell_dyn (int sector, int step) |
| Calling function for the cell_dyn** dynamic ecological modules. | |
| void | cell_dyn1 (int step) |
| Global dynamics and forcings. | |
| void | cell_dyn2 (int step) |
| Algae/periphyton (vertical) dynamics. | |
| void | cell_dyn4 (int step) |
| Organic Soil (vertical) dynamics. | |
| void | horizFlow (int step) |
| Horizontal raster fluxes. | |
| void | cell_dyn7 (int step) |
| Hydrologic (vertical) dynamics. | |
| void | cell_dyn8 (int step) |
| Macrophyte (vertical) dynamics. | |
| void | cell_dyn9 (int step) |
| Phosphorus (vertical) dynamics. | |
| void | cell_dyn10 (int step) |
| Salt/tracer (vertical) dynamics. | |
| void | cell_dyn12 (int step) |
| FLOCculent organic matter (vertical) dynamics. | |
| void | cell_dyn13 (int step) |
| Net settling of TP loss (stand-alone). | |
| float | tempCF (int form, float c1, float topt, float tmin, float tmax, float tempC) |
| Temperature control function for biology. | |
| void | init_static_data (void) |
| Initialize static spatial data. | |
| void | init_dynam_data (void) |
| Initialize dynamic spatial data. | |
| void | init_eqns (void) |
| Initialization of the model equations. | |
| void | init_canals (int irun) |
| Call to initialize the water managment canal network topology and data. | |
| void | init_succession (void) |
| Call to initialize the habitat succession module. | |
| void | reinitBIR (void) |
| Calls to re-initialize Basin/Indicator-Region data. | |
| void | reinitCanals (void) |
| Call to re-initialize canal storages. | |
| void | gen_output (int step, ViewParm *view) |
| Generate output. | |
| void | ReadGlobalParms (char *s_parm_name, int s_parm_relval) |
| Acquire the model parameters that are global to the domain. | |
| void | ReadHabParms (char *s_parm_name, int s_parm_relval) |
| Acquire the model parameters that are specific to different habitat types in the domain. | |
| void | ReadModExperimParms (char *s_parm_name, int s_parm_relval) |
| Acquire the model parameters that are used in special model experiments - used only in special, research-oriented applications. (Added for v2.6). NOTE that this was quickly set up for the ELM Peer Review (2006) project, and (at least in v2.6.0) should be used in conjunction with the spatial interpolators for rain and pET inputs. (because this is intended for scales that are potentially incompatible w/ simultaneous use of gridIO inputs on SFWMM stage/depth that are expected w/ gridIO rain/pET). NOTE: this has not been specifically developed/tested for inclusion within the automated sensitivity analyses - thus only been used at this point for the ModExperimParms_NOM nominal parameter set (Nov 2006). . | |
| void | PtInterp_read (void) |
| On-the-fly interpolations to develop spatial time series from point data. . | |
| void | get_map_dims (void) |
| Get the map dimensions of the global model array. | |
| void | alloc_memory () |
| Allocate memory. | |
| float | graph7 (unsigned char y, float x) |
| Time series interplator, data set 7. | |
| float | graph8 (unsigned char y, float x) |
| Time series interplator, data set 8. | |
| void | HabSwitch_Init (void) |
| Transfers habitat-specific parameters into data struct, allocate memory. | |
| unsigned char | HabSwitch (int ix, int iy, float *Water, float *Nutrient, int *Fire, float *Salinity, unsigned char *HAB) |
| Switches habitats (the option that is currently used). | |
| void | Run_Canal_Network (float *SWH, float *ElevMap, float *MC, float *GWV, float *poros, float *GWcond, double *NA, double *PA, double *SA, double *GNA, double *GPA, double *GSA, float *Unsat, float *sp_yield) |
| Runs the water management network. | |
| void | Canal_Network_Init (float baseDatum, float *elev) |
| Initialize the water managment network topology. | |
| void | CanalReInit () |
| Re-initialize the depths and concentrations in canals under the Positional Analysis mode. | |
| void | getInt (FILE *inFile, const char *lString, int *iValPtr) |
| Get an integer following a specific string. | |
| void | read_map_dims (const char *filename) |
| Establish the number of rows and columns of the model. | |
| void | write_output (int index, ViewParm *vp, void *Map, const char *filename, char Mtype, int step) |
| Determine which functions to call for model output. | |
| void | getChar (FILE *inFile, const char *lString, char *cValPtr) |
| Get a character following a specific string. | |
| void | init_pvar (VOIDP Map, UCHAR *mask, unsigned char Mtype, float iv) |
| Initialize a variable to a value. | |
| int | read_map_file (const char *filename, VOIDP Map, unsigned char Mtype, float scale_value, float offset_value) |
| Get the file and read/convert the data of map (array). | |
| int | scan_forward (FILE *infile, const char *tstring) |
| Scan forward until a particular string is found. | |
| VOIDP | nalloc (unsigned mem_size, const char var_name[]) |
| Allocate memory for a variable. | |
| float | FMOD (float x, float y) |
| Modulus of a pair of (double) arguments. | |
| void | PTSL_ReadLists (PTSeriesList *thisStruct, const char *ptsFileName, int index, float *timeStep, int *nPtTS, int col) |
| Point Time Series interpolation (unused): read raw point data. | |
| void | PTSL_CreatePointMap (PTSeriesList *pList, void *Map, unsigned char Mtype, int step, float scale) |
| Point Time Series interpolation (unused): generate interpolated spatial (map) data. | |
| void | stats (int step) |
| Calling function for budget and summary stats. | |
| void | alloc_mem_stats (void) |
| Allocate memory for the BIR-based and cell-based stats variables. | |
| void | BIRinit (void) |
| Set up the Basin & Indicator Region (BIR) linkages/inheritances. | |
| void | BIRoutfiles (void) |
| Open files and create headers for BIR output. | |
| void | BIRstats_reset (void) |
| Reset BIR (non-budget) statistics summations to zero. | |
| void | BIRbudg_reset (void) |
| Reset BIR budget summations to zero. | |
| void | Cell_reset_avg (void) |
| Zero the arrays holding selected variable averages in cells (after printing)/. | |
| void | Cell_reset_hydper (void) |
| Zero the array holding hydroperiod data in cells (after printing). | |
| void | Flux_SWater (int it, float *SURFACE_WAT, float *SED_ELEV, float *HYD_MANNINGS_N, double *STUF1, double *STUF2, double *STUF3, float *SfWat_vel_day) |
| Surface water horizontal flux. | |
| void | Flux_GWater (int it, float *SatWat, float *Unsat, float *SfWat, float *rate, float *poros, float *sp_yield, float *elev, double *gwSTUF1, double *gwSTUF2, double *gwSTUF3, double *swSTUF1, double *swSTUF2, double *swSTUF3) |
| Groundwater fluxing routine. | |
| float * | get_hab_parm (char *s_parm_name, int s_parm_relval, char *parmName) |
| Read the input data file to get a habitat-specfic parameter array. | |
| float | get_global_parm (char *s_parm_name, int s_parm_relval, char *parmName) |
| Read the input data file to get a global parameter. | |
| float | get_modexperim_parm (char *s_parm_name, int s_parm_relval, char *parmName) |
| Read the input data file to get a model experiment parameter. | |
| int | stage_data_wmm (float *) |
| int | rain_data_wmm (float *) |
| int | evap_data_wmm (float *) |
Variables | |
| float | DAYJUL |
| float | LATRAD |
| float | SOLALPHA |
| float | SOLALTCORR |
| float | SOLBETA |
| float | SOLCOSDEC |
| float | SOLDEC |
| float | SOLDEC1 |
| float | SOLELEV_SINE |
| float | SOLRADATMOS |
| float | SOLRISSET_HA |
| float | SOLRISSET_HA1 |
| float | dispParm_scaled |
| float | GP_BC_SfWat_add |
| float | GP_BC_SfWat_addHi |
| float | GP_BC_SatWat_add |
| float | GP_BC_SfWat_targ |
| int | GP_BC_InputRow |
| float | GP_BC_Pconc |
| float | GP_BC_Sconc |
| float | g7 [11][2] |
| float | g8 [11][2] |
| int | CalMonOut |
| PTSeriesList * | pTSList6 |
| PTSeriesList * | pTSList5 |
| PTSeriesList * | pTSList4 |
| PTSeriesList * | pTSList3 |
| PTSeriesList * | pTSList2 |
| float | Timestep0 = 1.0 |
| float | TMod0 = 0 |
| int | NPtTS0 = 0 |
| int | TIndex0 = 0 |
| float | Timestep1 = 1.0 |
| float | TMod1 = 0 |
| int | NPtTS1 = 0 |
| int | TIndex1 = 0 |
| float | Timestep2 = 1.0 |
| float | TMod2 = 0 |
| int | NPtTS2 = 0 |
| int | TIndex2 = 0 |
| float | Timestep3 = 1.0 |
| float | TMod3 = 0 |
| int | NPtTS3 = 0 |
| int | TIndex3 = 0 |
| float | Timestep4 = 1.0 |
| float | TMod4 = 0 |
| int | NPtTS4 = 0 |
| int | TIndex4 = 0 |
| float | Timestep5 = 1.0 |
| float | TMod5 = 0 |
| int | NPtTS5 = 0 |
| int | TIndex5 = 0 |
| float | Timestep6 = 1.0 |
| float | TMod6 = 0 |
| int | NPtTS6 = 0 |
| int | TIndex6 = 0 |
| basnDef ** | basn_list |
| basnDef * | basins |
| int | WatMgmtOn |
| int | ESPmodeON |
| int | HabSwitchOn |
| int | avgPrint |
| char * | OutputPath |
| char * | ProjName |
Detailed Description
Header file for the ecological Unit Model.
This defines or declares variables & functions that are global to Unit_Mod.c.
Note: documented with Doxygen, which expects specific syntax within special comments.
The Everglades Landscape Model (ELM).
last updated: Oct 2007
Definition in file unitmod.h.
Function Documentation
| int call_cell_dyn | ( | int | sector, | |
| int | step | |||
| ) |
Calling function for the cell_dyn** dynamic ecological modules.
This function calls the cell_dyn dynamic modules as defined in the Driver.parm data file. Normal order for calling ELM modules(Sectors): 1 0 7 10 9 2 8 4 12 99.
S#0 hydro: cell-cell horiz (&canals if WaterManagement is on)
S#1 global forcings
S#2 algae/periphyton
S#4 DOM/DOP
S#7 hydro: vertical
S#8 macrophytes
S#9 phosphorus
S#10 salt
S#12 FLOC
S#13 TP net settling
S#99 mass balance, budget, avg, hydroperiod, etc calculations
- Returns:
- rv (=1)
- Parameters:
-
sector The number of the cell_dyn** module being called step The current iteration number
Definition at line 150 of file UnitMod.c.
References cell_dyn1(), cell_dyn10(), cell_dyn12(), cell_dyn13(), cell_dyn2(), cell_dyn4(), cell_dyn7(), cell_dyn8(), cell_dyn9(), horizFlow(), and stats().
00151 { 00152 int rv=0; 00153 00154 switch(sector) { 00155 00156 case 99: { stats(step); rv=1; } break; 00157 case 0: { horizFlow(step); rv=1; } break; 00158 case 1: { cell_dyn1(step); rv=1; } break; 00159 case 2: { cell_dyn2(step); rv=1; } break; 00160 case 4: { cell_dyn4(step); rv=1; } break; 00161 case 7: { cell_dyn7(step); rv=1; } break; 00162 case 8: { cell_dyn8(step); rv=1; } break; 00163 case 9: { cell_dyn9(step); rv=1; } break; 00164 case 10: { cell_dyn10(step); rv=1; } break; 00165 case 12: { cell_dyn12(step); rv=1; } break; 00166 case 13: { cell_dyn13(step); rv=1; } break; 00167 default: printf("Warning, undefined sector number:%d\n",sector); 00168 } 00169 return rv; 00170 }

| void cell_dyn1 | ( | int | step | ) |
Global dynamics and forcings.
Includes interpolation-generators, gridIO spatial data input, and habitat switching. This module is reduced in scope from v2.1 & previous, now basically only used to read coarse-grid gridIO input data OR on-the-fly interpolators, and call the habitat succession function.
- Parameters:
-
step The current iteration number
Definition at line 193 of file UnitMod.c.
References Arctan, BCmodel_sfwat, boundcond_depth, boundcond_ETp, boundcond_rain, Cos, DAYJUL, debug, DT, evap_data_wmm(), Exp, FIREdummy, FMOD(), GP_ALTIT, HAB, HabSwitch(), HabSwitchOn, HP_HYD_POROSITY, HydRelDepPosNeg, HydTotHd, isPTSL, IsSubstituteModel, LATRAD, Max, msgStr, NPtTS2, NPtTS3, ON_MAP, OutputPath, PI, ProjName, PTSL_CreatePointMap(), pTSList2, pTSList3, rain_data_wmm(), s0, s1, SAL_SF_WT, SAT_WATER, SAT_WT_HEAD, SED_ELEV, SimTime, Sin, SOLALTCORR, SOLCOSDEC, SOLDEC, SOLDEC1, SOLELEV_SINE, SOLRADATMOS, SOLRISSET_HA, SOLRISSET_HA1, stage_data_wmm(), SURFACE_WAT, T, Tan, simTime::TIME, Timestep2, Timestep3, TPtoSOIL, UNSAT_DEPTH, UNSAT_WATER, and WriteMsg().
Referenced by call_cell_dyn().
00194 { 00195 int fail = -1; 00196 int ix, iy, cellLoc, stat=1; 00197 float fTemp; 00198 extern IsSubstituteModel; /* from generic_driver.h */ 00199 00200 /* v2.5 may be temporary? */ 00201 FILE *boundaryFile; 00202 char filename[40]; 00203 00204 /* SimTime.TIME is a (float) julian day counter since initialization (calc'd in main function, Generic_Driver.c source). 00205 (SimTime.TIME includes fractions of days if the vertical DT<1.0, but it is 00206 unlikely that the vertical DT will deviate from 1 day). */ 00207 00208 if ( debug > 4 ) { 00209 if (SimTime.TIME>60) printf("\nDebug level is very high - you're probably getting too much output data in %s/%s/Output/Debug/! Decrease your simulation length.\n", OutputPath, ProjName); 00210 exit(-1); 00211 } 00212 00213 /* v2.6 PTSL */ 00214 /* a choice of rainfall and pET input data routines */ 00215 if (!isPTSL) { /* do the gridIO inputs */ 00216 if (FMOD(SimTime.TIME,1.0) < 0.001) { /* daily re-mapping of coarse-grid input data */ 00217 /* remap sfwmm (or other grid) rain data to ELM grid scale */ 00218 stat=rain_data_wmm(boundcond_rain); 00219 if(stat==fail) 00220 { 00221 sprintf(msgStr,"Problem with rainfall data, time = %f.\n",SimTime.TIME); 00222 WriteMsg(msgStr,1); 00223 exit(fail); 00224 } 00225 00226 /* remap sfwmm (or other grid) potential ET data to ELM grid scale */ 00227 stat=evap_data_wmm(boundcond_ETp); 00228 if(stat==-1) 00229 { 00230 sprintf(msgStr,"Problem with ETp data, time = %f.\n",SimTime.TIME); 00231 WriteMsg(msgStr,1); 00232 exit(fail); 00233 } 00234 00235 /* remap sfwmm (or other grid) stage/water depth data to ELM grid scale */ 00236 stat=stage_data_wmm(boundcond_depth); 00237 if(stat==-1) 00238 { 00239 sprintf(msgStr,"Problem with stage data, time = %f.\n",SimTime.TIME); 00240 WriteMsg(msgStr,1); 00241 exit(fail); 00242 } 00243 00244 00245 for ( ix = 1; ix <= s0 ; ix++ ) 00246 for ( iy = 1 ; iy <= s1 ; iy++ ) 00247 { 00248 cellLoc= T(ix,iy); 00249 00250 /* v2.7.1 assign the external boundary condition model's depths to an ELM variable within the rectangular 00251 ELM domain (i.e., on-map and off-map), for output only (this variable not used in any calcs) */ 00252 if (IsSubstituteModel == 0) BCmodel_sfwat[cellLoc] = (boundcond_depth[cellLoc] > 0.0) ? (boundcond_depth[cellLoc]) : (0.0) ; 00253 00254 else { /* IsSubstituteModel is true */ 00255 if(ON_MAP[cellLoc]) { 00256 /* v2.8 - added feature to replace all ELM hydrologic calcs, substituting the Boundary Condition Model's 00257 "stage_minus_landElevation" variable into ELM hydro variables. 00258 This was done in order to post-process the SFWMM or NSM using the same procedures as ELM 00259 At initial setup, it was determined whether the user is requesting to only run cell_dyn1 (and stats, case #99) in UnitMod.c, 00260 which would make the IsSubstituteModel flag True */ 00261 00262 SURFACE_WAT[cellLoc] = (boundcond_depth[cellLoc] > 0.0) ? (boundcond_depth[cellLoc]) : (0.0) ; 00263 SAT_WT_HEAD[cellLoc] = (boundcond_depth[cellLoc] <= 0.0) ? (SED_ELEV[cellLoc] + boundcond_depth[cellLoc]) : (SED_ELEV[cellLoc]) ; 00264 HydTotHd[cellLoc] = SAT_WT_HEAD[cellLoc]+SURFACE_WAT[cellLoc]; 00265 SAT_WATER[cellLoc] = SAT_WT_HEAD[cellLoc] * HP_HYD_POROSITY[HAB[cellLoc]]; /* this isn't needed for current purposes, but may be of interest later */ 00266 UNSAT_DEPTH[cellLoc] = Max(SED_ELEV[cellLoc]-SAT_WT_HEAD[cellLoc],0.0); 00267 UNSAT_WATER[cellLoc] = UNSAT_DEPTH[cellLoc] * HP_HYD_POROSITY[HAB[cellLoc]] * 0.0; /* assume there is no moisture in pore spaces, thus never any unsat storage */ 00268 /* v2.8.2 new variable, for reporting purposes only: positive/negative water depth relative to land elevation (stage minus land elevation) */ 00269 HydRelDepPosNeg[cellLoc] = HydTotHd[cellLoc] - SED_ELEV[cellLoc]; 00270 } 00271 } 00272 00273 } 00274 00275 00276 00277 } /* end of mapping multi-grid, gridIO data */ 00278 } /* end of !isPTSL */ 00279 00280 else { /* instead of gridIO spatial data, calculate on-the-fly spatial interpolations of rain and pET point time series data */ 00281 /* NOTE that this option does not use gridIO stage/depth boundary condition data, and thus should be used in conjunction 00282 with the ModExperimParms_NOM synthetic boundary condition data */ 00283 fTemp = FMOD(SimTime.TIME,Timestep2*NPtTS2); 00284 PTSL_CreatePointMap(pTSList2,boundcond_rain,'f',(int)(fTemp/Timestep2),1.0); 00285 fTemp = FMOD(SimTime.TIME,Timestep3*NPtTS3); 00286 PTSL_CreatePointMap(pTSList3,boundcond_ETp,'f',(int)(fTemp/Timestep3),1.0); 00287 } 00288 00289 00290 00291 /* the "julian" day counter within a 365 day "year" */ 00292 DAYJUL = ( FMOD(SimTime.TIME,365.0) >0.0 ) ? ( FMOD(SimTime.TIME,365.0) ) : ( 365.0); 00293 /* DAYLENGTH is not used */ 00294 /* DAYLENGTH = AMPL*Sin((DAYJUL-79.0)*0.01721)+12.0; */ /* length of daylight (hours) */ 00295 00296 00297 /* Nikolov & Zeller (1992) solar radiation algorithm 00298 the algorithm and all parameters are published in the (Ecol. Mod., 61:149-168) manuscript, 00299 and the only modifiable parameters (in ELM database system) are local latitude and altitude */ 00300 SOLDEC1 = 0.39785*Sin(4.868961+0.017203*DAYJUL 00301 +0.033446*Sin(6.224111+0.017202*DAYJUL)); 00302 SOLCOSDEC = sqrt(1.0-SOLDEC1*SOLDEC1); 00303 SOLELEV_SINE = Sin(LATRAD)*SOLDEC1+Cos(LATRAD)*SOLCOSDEC; 00304 SOLALTCORR = (1.0-Exp(-0.014*(GP_ALTIT-274.0)/(SOLELEV_SINE*274.0))); 00305 SOLDEC = Arctan(SOLDEC1/sqrt(1.0-SOLDEC1*SOLDEC1)); 00306 SOLRISSET_HA1 = -Tan(LATRAD)*Tan(SOLDEC); 00307 SOLRISSET_HA = ( (SOLRISSET_HA1==0.0) ) ? ( PI*0.5 ) : ( ( (SOLRISSET_HA1<0.0) ) ? 00308 ( PI+Arctan(sqrt(1.0-SOLRISSET_HA1*SOLRISSET_HA1)/SOLRISSET_HA1) ) : 00309 ( Arctan(sqrt(1.0-SOLRISSET_HA1*SOLRISSET_HA1)/SOLRISSET_HA1))); 00310 SOLRADATMOS = 458.37*2.0*(1.0+0.033*Cos(360.0/365.0*PI/180.0*DAYJUL)) 00311 * ( Cos(LATRAD)*Cos(SOLDEC)*Sin(SOLRISSET_HA) 00312 + SOLRISSET_HA*180.0/(57.296*PI)*Sin(LATRAD)*Sin(SOLDEC)); 00313 00314 00315 /* daily habitat switching */ 00316 if ( (SimTime.TIME - (int)SimTime.TIME) < DT/2 && HabSwitchOn ) /* HabSwitchOn flag to invoke switching */ 00317 for(ix=1; ix<=s0; ix++) 00318 for(iy=1; iy<=s1; iy++) 00319 if(ON_MAP[cellLoc= T(ix,iy)]) { /* TPtoSoil == kg/kg */ 00320 HAB [cellLoc] = HabSwitch (ix, iy, SURFACE_WAT, TPtoSOIL, (int*)FIREdummy, SAL_SF_WT, HAB); 00321 } 00322 }
| void cell_dyn2 | ( | int | step | ) |
Algae/periphyton (vertical) dynamics.
Temporal dynamics of carbon and phosphorus of two communities of periphyton.
- Parameters:
-
step The current iteration number
Definition at line 344 of file UnitMod.c.
References ALG_INCID_LIGHT, ALG_LIGHT_CF, ALG_LIGHT_EXTINCT, ALG_REFUGE, ALG_SAT, ALG_TEMP_CF, ALG_TOT, ALG_WAT_CF, C_ALG, C_ALG_AVAIL_MORT, C_ALG_GPP, C_ALG_GPP_P, C_ALG_MORT, C_ALG_MORT_P, C_ALG_MORT_POT, C_ALG_NPP, C_ALG_NUT_CF, C_ALG_P, C_ALG_PC, C_ALG_PCrep, C_ALG_PROD_CF, C_ALG_RESP, C_ALG_RESP_POT, CELL_SIZE, conv_kgTOg, conv_kgTOmg, debug, DT, dynERRORnum, Exp, GP_alg_alkP_min, GP_alg_light_ext_coef, GP_ALG_LIGHT_SAT, GP_ALG_PC, GP_alg_R_accel, GP_ALG_RC_MORT, GP_ALG_RC_MORT_DRY, GP_ALG_RC_PROD, GP_ALG_RC_RESP, GP_ALG_REF_MULT, GP_ALG_SHADE_FACTOR, GP_ALG_TEMP_OPT, GP_alg_uptake_coef, GP_AlgComp, GP_algMortDepth, GP_C_ALG_KS_P, GP_C_ALG_threshTP, GP_MinCheck, GP_NC_ALG_KS_P, GP_PO4toTP, GP_PO4toTPint, GP_WQualMonitZ, H2O_TEMP, HAB, HP_ALG_MAX, MAC_LAI, Max, Min, msgStr, NC_ALG, NC_ALG_AVAIL_MORT, NC_ALG_GPP, NC_ALG_GPP_P, NC_ALG_MORT, NC_ALG_MORT_P, NC_ALG_MORT_POT, NC_ALG_NPP, NC_ALG_NUT_CF, NC_ALG_P, NC_ALG_PC, NC_ALG_PCrep, NC_ALG_PROD_CF, NC_ALG_RESP, NC_ALG_RESP_POT, ON_MAP, ramp, s0, s1, SFWT_VOL, SimTime, SOLRADGRD, SURFACE_WAT, T, tempCF(), simTime::TIME, TP_SF_WT, TP_SFWT_CONC, TP_SFWT_CONC_MG, TP_SFWT_UPTAK, True, UNSAT_DEPTH, and WriteMsg().
Referenced by call_cell_dyn().
00345 { 00346 int ix, iy, cellLoc; 00347 float reduc, min_litTemp,I_ISat, Z_extinct; 00348 double PO4Pconc, PO4P; 00349 float C_ALG_thresh_CF, mortPot; 00350 00351 for(ix=1; ix<=s0; ix++) { 00352 for(iy=1; iy<=s1; iy++) { 00353 00354 if(ON_MAP[cellLoc= T(ix,iy)]) { 00355 00356 /* these thresholds need updating when a habitat type of a grid cell changes */ 00357 ALG_REFUGE[cellLoc] = HP_ALG_MAX[HAB[cellLoc]]*GP_ALG_REF_MULT; 00358 ALG_SAT[cellLoc] = HP_ALG_MAX[HAB[cellLoc]]*0.9; 00359 00360 NC_ALG_AVAIL_MORT[cellLoc] = Max(NC_ALG[cellLoc]-ALG_REFUGE[cellLoc],0); 00361 C_ALG_AVAIL_MORT[cellLoc] = Max(C_ALG[cellLoc]-ALG_REFUGE[cellLoc],0); 00362 /* bio-avail P (PO4) is calc'd from TP, using pre-processed regression for predicting PO4 from TP */ 00363 /* assume that periphyton (microbial) alkaline phosphotase activity keeps PO4 at least 10% of TP conc */ 00364 PO4Pconc = Max(TP_SFWT_CONC_MG[cellLoc]*GP_PO4toTP + GP_PO4toTPint, 00365 0.10 * TP_SFWT_CONC_MG[cellLoc]); /* mg/L */ 00366 00367 /* light, water, temperature controls apply to both calc and non-calc */ 00368 ALG_LIGHT_EXTINCT[cellLoc] = GP_alg_light_ext_coef; 00369 /* algal self-shading implicit in density-dependent constraint function later */ 00370 ALG_INCID_LIGHT[cellLoc] = SOLRADGRD[cellLoc]*Exp(-MAC_LAI[cellLoc]*GP_ALG_SHADE_FACTOR); 00371 Z_extinct = SURFACE_WAT[cellLoc]*ALG_LIGHT_EXTINCT[cellLoc]; 00372 I_ISat = ALG_INCID_LIGHT[cellLoc]/GP_ALG_LIGHT_SAT; 00373 /* averaged over whole water column (based on Steele '65) */ 00374 ALG_LIGHT_CF[cellLoc] = ( Z_extinct > 0.0 ) ? 00375 ( 2.718/Z_extinct * (Exp(-I_ISat * Exp(-Z_extinct)) - Exp(-I_ISat)) ) : 00376 (I_ISat*Exp(1.0-I_ISat)); 00377 /* low-water growth constraint ready for something better based on data */ 00378 ALG_WAT_CF[cellLoc] = ( SURFACE_WAT[cellLoc]>0.0 ) ? ( 1.0 ) : ( 0.0); 00379 ALG_TEMP_CF[cellLoc] = tempCF(0, 0.20, GP_ALG_TEMP_OPT, 5.0, 40.0, H2O_TEMP[cellLoc]); 00380 00381 min_litTemp = Min(ALG_LIGHT_CF[cellLoc],ALG_TEMP_CF[cellLoc]); 00382 00383 /* the 2 communities have same form of growth response to avail phosphorus */ 00384 NC_ALG_NUT_CF[cellLoc] = 00385 Exp(-GP_alg_uptake_coef * Max(GP_NC_ALG_KS_P-PO4Pconc, 0.0)/GP_NC_ALG_KS_P) ; /* mg/L */ 00386 C_ALG_NUT_CF[cellLoc] = 00387 Exp(-GP_alg_uptake_coef * Max(GP_C_ALG_KS_P-PO4Pconc, 0.0)/GP_C_ALG_KS_P) ; /* mg/L */ 00388 00389 /* the form of the control function assumes that at very low 00390 P conc, the alkaline phosphotase activity of the microbial assemblage scavenges P, maintaining a minimum nutrient availability to community */ 00391 NC_ALG_PROD_CF[cellLoc] = Min(min_litTemp,ALG_WAT_CF[cellLoc])*Max(NC_ALG_NUT_CF[cellLoc], GP_alg_alkP_min); 00392 C_ALG_PROD_CF[cellLoc] = Min(min_litTemp,ALG_WAT_CF[cellLoc])*Max(C_ALG_NUT_CF[cellLoc], GP_alg_alkP_min); 00393 00394 NC_ALG_RESP_POT[cellLoc] = 00395 ( UNSAT_DEPTH[cellLoc]>GP_algMortDepth ) ? 00396 ( 0.0) : 00397 ( GP_ALG_RC_RESP*ALG_TEMP_CF[cellLoc]*NC_ALG_AVAIL_MORT[cellLoc] ); 00398 C_ALG_RESP_POT[cellLoc] = 00399 ( UNSAT_DEPTH[cellLoc]>GP_algMortDepth ) ? 00400 ( 0.0) : 00401 ( GP_ALG_RC_RESP*ALG_TEMP_CF[cellLoc] *C_ALG_AVAIL_MORT[cellLoc] ); 00402 00403 NC_ALG_RESP[cellLoc] = 00404 ( NC_ALG_RESP_POT[cellLoc]*DT>NC_ALG_AVAIL_MORT[cellLoc] ) ? 00405 ( NC_ALG_AVAIL_MORT[cellLoc]/DT ) : 00406 ( NC_ALG_RESP_POT[cellLoc]); 00407 C_ALG_RESP[cellLoc] = 00408 ( C_ALG_RESP_POT[cellLoc]*DT>C_ALG_AVAIL_MORT[cellLoc] ) ? 00409 ( C_ALG_AVAIL_MORT[cellLoc]/DT ) : 00410 ( C_ALG_RESP_POT[cellLoc]); 00411 00412 /* this is the threshold control function that increases 00413 calcareous/native periph mortality (likely due to loss of calcareous sheath) as P conc. increases */ 00414 C_ALG_thresh_CF = Min(exp(GP_alg_R_accel*Max( TP_SFWT_CONC_MG[cellLoc]-GP_C_ALG_threshTP,0.0)/GP_C_ALG_threshTP), 100.0); 00415 00416 NC_ALG_MORT_POT[cellLoc] = 00417 ( UNSAT_DEPTH[cellLoc]>GP_algMortDepth ) ? 00418 ( NC_ALG_AVAIL_MORT[cellLoc]*GP_ALG_RC_MORT_DRY ) : 00419 ( NC_ALG_AVAIL_MORT[cellLoc]*GP_ALG_RC_MORT); 00420 C_ALG_MORT_POT[cellLoc] = 00421 ( UNSAT_DEPTH[cellLoc]>GP_algMortDepth ) ? 00422 ( C_ALG_AVAIL_MORT[cellLoc]*GP_ALG_RC_MORT_DRY ) : 00423 ( C_ALG_thresh_CF * C_ALG_AVAIL_MORT[cellLoc]*GP_ALG_RC_MORT); 00424 00425 NC_ALG_MORT[cellLoc] = 00426 ( (NC_ALG_MORT_POT[cellLoc]+NC_ALG_RESP[cellLoc])*DT>NC_ALG_AVAIL_MORT[cellLoc] ) ? 00427 ( (NC_ALG_AVAIL_MORT[cellLoc]-NC_ALG_RESP[cellLoc]*DT)/DT ) : 00428 ( NC_ALG_MORT_POT[cellLoc]); 00429 C_ALG_MORT[cellLoc] = 00430 ( (C_ALG_MORT_POT[cellLoc]+C_ALG_RESP[cellLoc])*DT>C_ALG_AVAIL_MORT[cellLoc] ) ? 00431 ( (C_ALG_AVAIL_MORT[cellLoc]-C_ALG_RESP[cellLoc]*DT)/DT ) : 00432 ( C_ALG_MORT_POT[cellLoc]); 00433 00434 /* gross production of the 2 communities (gC/m2/d, NOT kgC/m2/d) */ 00435 /* density constraint on both noncalc and calc, competition effect accentuated by calc algae */ 00436 00437 NC_ALG_GPP[cellLoc] = NC_ALG_PROD_CF[cellLoc]*GP_ALG_RC_PROD*NC_ALG[cellLoc] 00438 *Max( (1.0-(GP_AlgComp*C_ALG[cellLoc]+NC_ALG[cellLoc])/HP_ALG_MAX[HAB[cellLoc]]),0.0); 00439 C_ALG_GPP[cellLoc] = C_ALG_PROD_CF[cellLoc]*GP_ALG_RC_PROD*C_ALG[cellLoc] 00440 *Max( (1.0-( C_ALG[cellLoc]+NC_ALG[cellLoc])/HP_ALG_MAX[HAB[cellLoc]]),0.0); 00441 00442 00443 /* P uptake is dependent on available P and is relative to a maximum P:C ratio for the tissue 00444 (g C/m^2/d * P:Cmax * dimless * dimless = gP/m2/d (NOT kg) )*/ 00445 NC_ALG_GPP_P[cellLoc] = NC_ALG_GPP[cellLoc] *GP_ALG_PC * NC_ALG_NUT_CF[cellLoc] 00446 * Max(1.0-NC_ALG_PC[cellLoc]/GP_ALG_PC, 0.0); 00447 C_ALG_GPP_P[cellLoc] = C_ALG_GPP[cellLoc] * GP_ALG_PC * C_ALG_NUT_CF[cellLoc] 00448 * Max(1.0-C_ALG_PC[cellLoc]/GP_ALG_PC, 0.0); 00449 00450 /* check for available P mass (the nutCF does not) */ 00451 PO4P = ramp(Min(PO4Pconc * SFWT_VOL[cellLoc], conv_kgTOg*TP_SF_WT[cellLoc]) ); /*g P available; v2.5 put in ramp (constrain non-neg) */ 00452 reduc = ( (NC_ALG_GPP_P[cellLoc]+C_ALG_GPP_P[cellLoc]) > 0) ? 00453 (PO4P / ( (NC_ALG_GPP_P[cellLoc]+C_ALG_GPP_P[cellLoc])*CELL_SIZE*DT) ) : 00454 (1.0); 00455 /* can have high conc, but low mass of P avail, in presence of high peri biomass and high demand */ 00456 /* reduce the production proportionally if excess demand is found */ 00457 if (reduc < 1.0) { 00458 NC_ALG_GPP[cellLoc] *= reduc; 00459 NC_ALG_GPP_P[cellLoc] *= reduc; 00460 C_ALG_GPP[cellLoc] *= reduc; 00461 C_ALG_GPP_P[cellLoc] *= reduc; 00462 } 00463 00464 /* state variables calc'd (gC/m2, NOT kgC/m2) */ 00465 NC_ALG[cellLoc] = NC_ALG[cellLoc] 00466 + (NC_ALG_GPP[cellLoc] 00467 - NC_ALG_RESP[cellLoc] - NC_ALG_MORT[cellLoc]) * DT; 00468 00469 C_ALG[cellLoc] = C_ALG[cellLoc] 00470 + (C_ALG_GPP[cellLoc] 00471 - C_ALG_RESP[cellLoc] - C_ALG_MORT[cellLoc]) * DT; 00472 00473 /* carbon NPP not currently used elsewhere, only for output */ 00474 NC_ALG_NPP[cellLoc] = NC_ALG_GPP[cellLoc]-NC_ALG_RESP[cellLoc]; 00475 C_ALG_NPP[cellLoc] = C_ALG_GPP[cellLoc]-C_ALG_RESP[cellLoc]; 00476 /* carbon total biomass of both communities, not currently used elsewhere, only for output */ 00477 ALG_TOT[cellLoc] = NC_ALG[cellLoc] + C_ALG[cellLoc]; 00478 00479 00480 /* now calc the P associated with the C fluxes (GPP_P already calc'd) */ 00481 mortPot = (double) NC_ALG_MORT[cellLoc] * NC_ALG_PC[cellLoc]; 00482 NC_ALG_MORT_P[cellLoc] = (mortPot*DT>NC_ALG_P[cellLoc]) ? 00483 (NC_ALG_P[cellLoc]/DT) : 00484 (mortPot); 00485 mortPot = (double) C_ALG_MORT[cellLoc] * C_ALG_PC[cellLoc]; 00486 C_ALG_MORT_P[cellLoc] = (mortPot*DT>C_ALG_P[cellLoc]) ? 00487 (C_ALG_P[cellLoc]/DT) : 00488 (mortPot); 00489 00490 00491 /* state variables calc'd (gP/m2, NOT kgP/m2) */ 00492 NC_ALG_P[cellLoc] = NC_ALG_P[cellLoc] 00493 + (NC_ALG_GPP_P[cellLoc] - NC_ALG_MORT_P[cellLoc]) * DT; 00494 00495 C_ALG_P[cellLoc] = C_ALG_P[cellLoc] 00496 + (C_ALG_GPP_P[cellLoc] - C_ALG_MORT_P[cellLoc]) * DT; 00497 00498 NC_ALG_PC[cellLoc] = (NC_ALG[cellLoc]>0.0) ? 00499 (NC_ALG_P[cellLoc]/ NC_ALG[cellLoc]) : 00500 ( GP_ALG_PC * 0.03); /* default to 3% of max P:C */ 00501 C_ALG_PC[cellLoc] = (C_ALG[cellLoc]>0.0) ? 00502 (C_ALG_P[cellLoc]/ C_ALG[cellLoc]) : 00503 ( GP_ALG_PC * 0.03 ); /* default to 3% of max P:C */ 00504 NC_ALG_PCrep[cellLoc] = (float)NC_ALG_PC[cellLoc] * conv_kgTOmg; /* variable for output _rep-orting only */ 00505 C_ALG_PCrep[cellLoc] = (float)C_ALG_PC[cellLoc] * conv_kgTOmg; /* variable for output _rep-orting only */ 00506 00507 if (debug > 0 && NC_ALG[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg NC_ALG C biomass (%f g/m2) in cell (%d,%d)!", 00508 SimTime.TIME, NC_ALG[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 00509 if (debug > 0 && C_ALG[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg C_ALG C biomass (%f g/m2) in cell (%d,%d)!", 00510 SimTime.TIME, C_ALG[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 00511 if (debug > 0 && NC_ALG_P[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg NC_ALG_P P biomass (%f g/m2) in cell (%d,%d)!", 00512 SimTime.TIME, NC_ALG_P[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 00513 if (debug > 0 && C_ALG_P[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg C_ALG_P P biomass (%f g/m2) in cell (%d,%d)!", 00514 SimTime.TIME, C_ALG_P[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 00515 00516 00517 TP_SFWT_UPTAK[cellLoc] = (NC_ALG_GPP_P[cellLoc]+C_ALG_GPP_P[cellLoc]) 00518 *0.001*CELL_SIZE; /* gP/m2 => kg P */ 00519 /* recalc P in surface water state variable (kg P) */ 00520 TP_SF_WT[cellLoc] = TP_SF_WT[cellLoc] - TP_SFWT_UPTAK[cellLoc] * DT; 00521 TP_SFWT_CONC[cellLoc] = 00522 ( SFWT_VOL[cellLoc] > 0.0 ) ? 00523 ( TP_SF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 00524 ( 0.0); /* used in P fluxes for mass balance */ 00525 TP_SFWT_CONC_MG[cellLoc] = 00526 ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? 00527 (TP_SFWT_CONC[cellLoc]*conv_kgTOg) : 00528 (0.0); /* (g/m3==mg/L) used for reporting and other modules to evaluate P conc when water is present */ 00529 00530 } 00531 } 00532 } 00533 }
| void cell_dyn4 | ( | int | step | ) |
Organic Soil (vertical) dynamics.
Temporal dynamics of Deposited Organic Matter and Deposited Organic Phosphorus.
- Parameters:
-
step The current iteration number
Definition at line 553 of file UnitMod.c.
References C_ALG_P, CELL_SIZE, conv_kgTOg, conv_kgTOmg, DEPOS_ORG_MAT, DIM, DOM_BD, DOM_DECOMP, DOM_DECOMP_POT, DOM_FR_FLOC, DOM_fr_nphBio, DOM_P_OM, DOM_QUALITY_CF, DOM_SED_AEROB_Z, DOM_SED_ANAEROB_Z, DOM_TEMP_CF, DOM_Z, DOP, DOP_DECOMP, DOP_FLOC, DOP_nphBio, DT, Exp, FLOC_DEPO, FlocP, FlocP_DEPO, GP_calibDecomp, GP_DOM_decomp_coef, GP_DOM_DECOMP_POPT, GP_DOM_DECOMP_TOPT, GP_DOM_DECOMPRED, GP_DOM_RCDECOMP, GP_WQualMonitZ, H2O_TEMP, HAB, HP_DOM_AEROBTHIN, HP_DOM_MAXDEPTH, HP_TP_CONC_GRAD, HYD_DOM_ACTWAT_PRES, HYD_DOM_ACTWAT_VOL, HYD_SED_WAT_VOL, Inorg_Z, mac_nph_P, mac_ph_P, Max, Min, NC_ALG_P, nphbio_mort_OM, nphbio_mort_P, ON_MAP, P_SUM_CELL, s0, s1, SED_ELEV, SED_INACT_Z, SFWT_VOL, soil_MOIST_CF, SURFACE_WAT, T, tempCF(), TP_Act_to_Tot, TP_Act_to_TotRep, TP_SED_CONC, TP_SED_WT, TP_SED_WT_AZ, TP_SEDWT_CONCACT, TP_SEDWT_CONCACTMG, TP_SF_WT, TP_SFWT_CONC, TP_SFWT_CONC_MG, TP_SORB, TPtoSOIL, TPtoSOIL_rep, TPtoVOL, TPtoVOL_rep, and UNSAT_DEPTH.
Referenced by call_cell_dyn().
00554 { 00555 int ix, iy, cellLoc; 00556 float TPsoil, TP_sedMin, TP_sfMin; 00557 00558 00559 00560 for(ix=1; ix<=s0; ix++) { 00561 for(iy=1; iy<=s1; iy++) { 00562 00563 if(ON_MAP[cellLoc= T(ix,iy)]) { 00564 00565 00566 /* inputs of organic matter (kg OM/m2)*/ 00567 DOM_fr_nphBio[cellLoc] = nphbio_mort_OM[cellLoc]; 00568 DOM_FR_FLOC[cellLoc] = FLOC_DEPO[cellLoc] ; 00569 00570 /* losses of organic matter (kg OM/m2) */ 00571 00572 DOM_QUALITY_CF[cellLoc] = 00573 Min(Exp(-GP_DOM_decomp_coef * Max(GP_DOM_DECOMP_POPT-TP_SEDWT_CONCACTMG[cellLoc], 0.0) 00574 /GP_DOM_DECOMP_POPT),1.0) ; /* mg/L */ 00575 DOM_TEMP_CF[cellLoc] = tempCF(0, 0.20, GP_DOM_DECOMP_TOPT, 5.0, 40.0, H2O_TEMP[cellLoc]); 00576 DOM_SED_AEROB_Z[cellLoc] = Min(Max(UNSAT_DEPTH[cellLoc],HP_DOM_AEROBTHIN[HAB[cellLoc]]),HP_DOM_MAXDEPTH[HAB[cellLoc]]); 00577 DOM_SED_ANAEROB_Z[cellLoc] = HP_DOM_MAXDEPTH[HAB[cellLoc]]-DOM_SED_AEROB_Z[cellLoc]; 00578 00579 /* GP_calibDecomp is an adjustable calib parm */ 00580 DOM_DECOMP_POT[cellLoc] = (double) GP_calibDecomp*GP_DOM_RCDECOMP*DOM_QUALITY_CF[cellLoc]*DOM_TEMP_CF[cellLoc]*DEPOS_ORG_MAT[cellLoc] 00581 *(Min(DOM_SED_AEROB_Z[cellLoc]/HP_DOM_MAXDEPTH[HAB[cellLoc]],1.0)*soil_MOIST_CF[cellLoc] 00582 +GP_DOM_DECOMPRED*Min(DOM_SED_ANAEROB_Z[cellLoc]/HP_DOM_MAXDEPTH[HAB[cellLoc]],1.0) ); 00583 DOM_DECOMP[cellLoc] = 00584 ( (DOM_DECOMP_POT[cellLoc])*DT>DEPOS_ORG_MAT[cellLoc] ) ? 00585 ( (DEPOS_ORG_MAT[cellLoc])/DT ) : 00586 ( DOM_DECOMP_POT[cellLoc]); 00587 /* calc state var (kg OM/m2) */ 00588 DEPOS_ORG_MAT[cellLoc] = DEPOS_ORG_MAT[cellLoc] + 00589 ( DOM_fr_nphBio[cellLoc] + DOM_FR_FLOC[cellLoc] 00590 - DOM_DECOMP[cellLoc] ) * DT; 00591 00592 /* soil elevation */ 00593 DOM_Z[cellLoc] = (double) DEPOS_ORG_MAT[cellLoc] / DOM_BD[cellLoc] ; /* (m) organic depth */ 00594 SED_ELEV[cellLoc] = DOM_Z[cellLoc]+Inorg_Z[cellLoc]+SED_INACT_Z[cellLoc]; /* total land surface elevation, including model GP_DATUM_DISTANCE below zero of land elev datum (NGVD/NAVD) (m) */ 00595 00596 /* P DOM stoich (kg P /m2) */ 00597 DOP_nphBio[cellLoc] = nphbio_mort_P[cellLoc]; 00598 DOP_FLOC[cellLoc] = FlocP_DEPO[cellLoc]; 00599 00600 DOP_DECOMP[cellLoc] = (double) DOM_DECOMP[cellLoc] * DOM_P_OM[cellLoc]; 00601 00602 /* calc state var of total "organic" P in soil (NOT including dissolved in pore water or sorbed) (kgP/m2) */ 00603 DOP[cellLoc] = DOP[cellLoc] 00604 + ( DOP_nphBio[cellLoc] + DOP_FLOC[cellLoc] 00605 - DOP_DECOMP[cellLoc]) * DT; /* kgP/m2 */ 00606 00607 /* now the P ratio */ 00608 DOM_P_OM[cellLoc] = (DEPOS_ORG_MAT[cellLoc]>0.0) ? ( DOP[cellLoc] / DEPOS_ORG_MAT[cellLoc]) : (0.0); /* kgP/kgOM */ 00609 TPsoil = DOP[cellLoc]*CELL_SIZE + TP_SORB[cellLoc]; /* kg TP in soil */ 00610 TPtoSOIL[cellLoc] = ((DEPOS_ORG_MAT[cellLoc]*CELL_SIZE + DIM[cellLoc])>0.0) ? 00611 ( TPsoil / (DEPOS_ORG_MAT[cellLoc]*CELL_SIZE + DIM[cellLoc]) ) : (0.0); /* kgP/kgsoil */ 00612 TPtoVOL[cellLoc] = (CELL_SIZE * DOM_Z[cellLoc]>0.0) ? 00613 (TPsoil / (CELL_SIZE * DOM_Z[cellLoc]) ) : 00614 (0.0); /* kgP/m3 soil */ 00615 00616 TPtoSOIL_rep[cellLoc] = TPtoSOIL[cellLoc] * conv_kgTOmg; /* reporting purposes only (kg/kg->mg/kg) */ 00617 TPtoVOL_rep[cellLoc] = TPtoVOL[cellLoc] * conv_kgTOg; /* reporting purposes only (kg/m3->g/m3 == ug/cm3) */ 00618 00619 /* now the P gain in sed water with decomp 00620 a small proportion goes into surface water P (below) */ 00621 TP_sedMin = (1.0 - HP_DOM_AEROBTHIN[HAB[cellLoc]] / HP_DOM_MAXDEPTH[HAB[cellLoc]] ) 00622 * DOP_DECOMP[cellLoc] * CELL_SIZE; 00623 00624 /* calc P in sed water state variables (kg P) */ 00625 TP_SED_WT[cellLoc] = TP_SED_WT[cellLoc] + TP_sedMin * DT; 00626 /* this is the active zone, where uptake, sorption, and mineralization take place */ 00627 TP_SED_WT_AZ[cellLoc] = TP_SED_WT_AZ[cellLoc] + TP_sedMin * DT; 00628 00629 TP_SED_CONC[cellLoc] = (HYD_SED_WAT_VOL[cellLoc]>0.0) ? 00630 (TP_SED_WT[cellLoc] / HYD_SED_WAT_VOL[cellLoc]) : 00631 (0.0); 00632 TP_SEDWT_CONCACT[cellLoc] = ( HYD_DOM_ACTWAT_PRES[cellLoc] > 0.0) ? 00633 ( TP_SED_WT_AZ[cellLoc]/HYD_DOM_ACTWAT_VOL[cellLoc] ) : 00634 (TP_SED_CONC[cellLoc]); /* g/L */ 00635 TP_SEDWT_CONCACTMG[cellLoc] = TP_SEDWT_CONCACT[cellLoc]*conv_kgTOg; /* g/m3==mg/L */ 00636 00637 /* now store the ratio of the conc in the active zone relative to total, prior to horiz fluxes 00638 ***** very simple constant, code in transition **** */ 00639 TP_Act_to_Tot[cellLoc] = 1.0 / HP_TP_CONC_GRAD[HAB[cellLoc]]; 00640 TP_Act_to_TotRep[cellLoc] = (float) TP_Act_to_Tot[cellLoc]; 00641 00642 /* now the P gain in surface water with decomp in the very thin upper layer of the soil */ 00643 /* if there is no surface water present, assume that this 00644 relative contribution will be an additional sorbed component that 00645 is introduced to surface water column immediately upon hydration 00646 with surface water */ 00647 TP_sfMin = HP_DOM_AEROBTHIN[HAB[cellLoc]] / HP_DOM_MAXDEPTH[HAB[cellLoc]] 00648 * DOP_DECOMP[cellLoc] * CELL_SIZE; 00649 00650 /* calc P in surface water state variable (kg P) */ 00651 TP_SF_WT[cellLoc] = TP_SF_WT[cellLoc] + TP_sfMin * DT; 00652 TP_SFWT_CONC[cellLoc] = 00653 ( SFWT_VOL[cellLoc] > 0.0 ) ? 00654 ( TP_SF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 00655 ( 0.0); /* used in P fluxes for mass balance */ 00656 TP_SFWT_CONC_MG[cellLoc] = 00657 ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? 00658 (TP_SFWT_CONC[cellLoc]*conv_kgTOg) : 00659 (0.0); /* (g/m3==mg/L) used for reporting and other modules to evaluate P conc when water is present */ 00660 00661 /* for reporting only: calc sum of all P storages in grid cells (budget calcs do same for Basin/Indicator-Regions) (g P /m^2) */ 00662 P_SUM_CELL[cellLoc] = ( (C_ALG_P[cellLoc] + NC_ALG_P[cellLoc]) * 0.001 * CELL_SIZE + /* gP/m2 => kgP */ 00663 (mac_nph_P[cellLoc] + mac_ph_P[cellLoc] )* CELL_SIZE + /* kgP/m2 => kgP */ 00664 TP_SORB[cellLoc] + 00665 ( FlocP[cellLoc] + DOP[cellLoc] ) * CELL_SIZE + /* kgP/m2 => kgP */ 00666 TP_SED_WT[cellLoc] + TP_SF_WT[cellLoc] ) /* kgP */ 00667 /CELL_SIZE * conv_kgTOg; /* kg P/m^2 => g P/m^2 */ 00668 00669 } 00670 } 00671 } 00672 }
| void horizFlow | ( | int | step | ) |
Horizontal raster fluxes.
Flows of water and dissolved/suspended constituents (phosphorus, salt/tracer) among cells and in water management network.
- Parameters:
-
step The current iteration number
Definition at line 682 of file UnitMod.c.
References DINdummy, Flux_GWater(), Flux_SWater(), HP_HYD_POROSITY, HP_HYD_SPEC_YIELD, hyd_iter, HYD_MANNINGS_N, HYD_RCCONDUCT, msgStr, ON_MAP, Run_Canal_Network(), s0, s1, SALT_SED_WT, SALT_SURF_WT, SAT_WATER, SED_ELEV, SF_WT_VEL_mag, SURFACE_WAT, T, TP_SED_WT, TP_SF_WT, UNSAT_WATER, usrErr0(), and WatMgmtOn.
Referenced by call_cell_dyn().
00683 { 00684 int it; 00685 int ix, iy; 00686 00687 /* v2.7.0 adding velocity output capabilities */ 00688 for(ix=1; ix<=s0; ix++) { 00689 for(iy=1; iy<=s1; iy++) { 00690 if(ON_MAP[T(ix,iy)]) SF_WT_VEL_mag[T(ix,iy)]=0.0; 00691 } 00692 } 00693 00694 for ( it = 0; it < hyd_iter; it++ ) 00695 { 00696 sprintf(msgStr,"\b\b\b%3d",it); 00697 usrErr0(msgStr); 00698 00699 /* horizontal raster-vector canal fluxes and water management functions 00700 Water Management switch set at runtime in Driver.parm 00701 this routine also integrates surface, unsaturated, and saturated exchanges 00702 */ 00703 /* Nitrogen (DINdummy) argument is dummy argument placeholder */ 00704 if (WatMgmtOn ) { 00705 Run_Canal_Network(SURFACE_WAT,SED_ELEV,HYD_MANNINGS_N,SAT_WATER,HP_HYD_POROSITY, 00706 HYD_RCCONDUCT, DINdummy,TP_SF_WT,SALT_SURF_WT,DINdummy,TP_SED_WT,SALT_SED_WT, 00707 UNSAT_WATER,HP_HYD_SPEC_YIELD ); 00708 } 00709 00710 00711 /* Function for horiz fluxing of surface water, no exchange with sat/unsat water */ 00712 /* if the order of the solute is changed, be sure to change the mass bal info in FluxSWstuff fcn */ 00713 /* Nitrogen (DINdummy) argument is dummy argument placeholder */ 00714 Flux_SWater(it,SURFACE_WAT,SED_ELEV,HYD_MANNINGS_N,SALT_SURF_WT,DINdummy,TP_SF_WT,SF_WT_VEL_mag); 00715 00716 /* Function for horiz fluxing of ground water and its vertical itegration with unsat and surface water 00717 It is only called every other hyd_iter iteration, and passes the realized number of gwat iterations to the function. 00718 If the order of the solutes is changed, be sure to change the mass bal info in FluxGWstuff fcn 00719 */ 00720 /* Nitrogen (DINdummy) argument is dummy argument placeholder */ 00721 if ( it%2 ) 00722 Flux_GWater((it-1)/2, SAT_WATER, UNSAT_WATER, SURFACE_WAT, HYD_RCCONDUCT, HP_HYD_POROSITY, 00723 HP_HYD_SPEC_YIELD, SED_ELEV, SALT_SED_WT, DINdummy, TP_SED_WT, SALT_SURF_WT, DINdummy, TP_SF_WT); 00724 00725 } /* end of hyd_iter */ 00726 00727 /* v2.7.0 adding velocity output capabilities */ 00728 for(ix=1; ix<=s0; ix++) { 00729 for(iy=1; iy<=s1; iy++) { 00730 SF_WT_VEL_mag[T(ix,iy)]= SF_WT_VEL_mag[T(ix,iy)] / hyd_iter / 2.0; /* v2.8.4 - no changes, but explored the /hyd_iter/2.0 etc */ 00731 } 00732 } 00733 00734 }
| void cell_dyn7 | ( | int | step | ) |
Hydrologic (vertical) dynamics.
Temporal dynamics of water storages (not including horizontal flows).
- Parameters:
-
step The current iteration number
- Remarks:
- The horizontal solutions (in horizFlow function) makes other vertical calcs to integrate the three vertical zones of water storage.
Definition at line 758 of file UnitMod.c.
References Abs, AIR_TEMP, boundcond_ETp, boundcond_rain, CELL_SIZE, debug, DT, dynERRORnum, Exp, GP_calibET, GP_DetentZ, GP_HYD_RCRECHG, GP_mann_height_coef, GP_SOLOMEGA, graph7(), graph8(), H2O_TEMP, HAB, HP_DOM_MAXDEPTH, HP_HYD_POROSITY, HP_HYD_RCINFILT, HP_HYD_SPEC_YIELD, HP_MAC_MAXROUGH, HP_MAC_MINROUGH, HP_NPHBIO_ROOTDEPTH, HYD_DOM_ACTWAT_PRES, HYD_DOM_ACTWAT_VOL, HYD_ET, HYD_EVAP_CALC, HYD_MANNINGS_N, HYD_SAT_POT_TRANS, HYD_SED_WAT_VOL, HYD_TOT_POT_TRANSP, HYD_TRANSP, HYD_UNSAT_POT_TRANS, HYD_WATER_AVAIL, HydRelDepPosNeg, HydTotHd, LAI_eff, MAC_HEIGHT, MAC_LAI, MAC_REL_BIOM, MAC_WATER_AVAIL_CF, Max, Min, msgStr, ON_MAP, s0, s1, SAT_TO_UNSAT_FL, SAT_VS_UNSAT, SAT_WATER, SAT_WT_HEAD, SAT_WT_RECHG, SAT_WT_TRANSP, SED_ELEV, SF_WT_EVAP, SF_WT_FROM_RAIN, SF_WT_INFILTRATION, SF_WT_POT_INF, SF_WT_TO_SAT_DOWNFLOW, SFWT_VOL, SimTime, SOLALPHA, SOLALTCORR, SOLBETA, SOLRAD274, SOLRADATMOS, SOLRADGRD, SURFACE_WAT, T, simTime::TIME, True, UNSAT_AVAIL, UNSAT_CAP, UNSAT_DEPTH, UNSAT_HYD_COND_CF, UNSAT_MOIST_PRP, UNSAT_PERC, UNSAT_TO_SAT_FL, UNSAT_TRANSP, UNSAT_WATER, UNSAT_WT_POT, and WriteMsg().
Referenced by call_cell_dyn().
00759 { 00760 int ix, iy, cellLoc; 00761 float SatWat_Root_CF, field_cap; 00762 float mann_height, N_density, f_LAI_eff, sfwat_pr1; 00763 float cloudy=0.0; 00764 00765 /* the horizontal raster and vector fluxes are always called before this cell_dyn */ 00766 for(ix=1; ix<=s0; ix++) { 00767 for(iy=1; iy<=s1; iy++) { 00768 if(ON_MAP[cellLoc= T(ix,iy)]) { 00769 00770 if (debug > 3) { /* these are old, relatively coarse checks - surface-ground water integration occurs in Fluxes.c */ 00771 if (SAT_WT_HEAD[cellLoc] - 0.01 > SED_ELEV[cellLoc] ) { 00772 sprintf(msgStr,"Day %6.1f. Warning - SAT_WT_HEAD exceeds elev in cell (%d,%d) by %f.", 00773 SimTime.TIME, ix,iy,(SAT_WT_HEAD[cellLoc] - SED_ELEV[cellLoc]) ); 00774 WriteMsg( msgStr,True ); } 00775 if (SURFACE_WAT[cellLoc] > 0.2 && UNSAT_DEPTH[cellLoc] > 0.1 ) { 00776 sprintf(msgStr,"Day: %6.1f. Warning - large sfwat depth (%5.2f m) in presence of unsat= %5.2f m, %4.2f %% moist, in cell (%d,%d).", 00777 SimTime.TIME, SURFACE_WAT[cellLoc], UNSAT_DEPTH[cellLoc],UNSAT_MOIST_PRP[cellLoc], ix,iy ); 00778 WriteMsg( msgStr,True ); } 00779 if (SAT_WATER[cellLoc] < -0.01) { /* this seems unnecessary but... */ 00780 sprintf(msgStr,"Day %6.1f: capacityERR - neg SAT_WATER (%f m) in cell (%d,%d) before cell_dyn7!", 00781 SimTime.TIME, SAT_WATER[cellLoc], ix,iy ); 00782 WriteMsg( msgStr,True ); dynERRORnum++; } 00783 if (SURFACE_WAT[cellLoc] < -0.01) { 00784 sprintf(msgStr,"Day %6.1f: capacityERR - neg SURFACE_WAT (%f m) in cell (%d,%d) before cell_dyn7!", 00785 SimTime.TIME, SURFACE_WAT[cellLoc], ix,iy ); 00786 WriteMsg( msgStr,True ); dynERRORnum++; } 00787 } 00788 00789 /* note that rainfall during a time step is added to surface water storage and available */ 00790 /* for runoff (horizFlow) before the calc of infiltration & ET associated with that new input */ 00791 /* (infiltration/ET etc will be of avail water the next time step after a rainfall event and horiz flows) */ 00792 /* NSM/SFWMM rainfall input data, created in cell_dyn1, convert here from tenths of mm to meters */ 00793 SF_WT_FROM_RAIN[cellLoc] = boundcond_rain[cellLoc]*0.0001; /* tenths of mm *0.0001 = m */ 00794 00795 /* solar radiation at altitude of 274m in atmosphere cal/cm2/d) */ 00796 /* v2.2+, CLOUDY (cloudiness) spatiotemporal data "temporarily" unavailable, is constant in space and time, local var "cloudy" */ 00797 SOLRAD274[cellLoc] = SOLRADATMOS*(SOLBETA-GP_SOLOMEGA* ( ( cloudy>0.0 ) ? ( cloudy ) : ( 0.0) ) ) -SOLALPHA; 00798 SOLRADGRD[cellLoc] = SOLRAD274[cellLoc]+((SOLRADATMOS+1.0)-SOLRAD274[cellLoc])*SOLALTCORR; 00799 H2O_TEMP[cellLoc] = AIR_TEMP[cellLoc]; /* v2.2+, temperature data "temporarily" unavailable, is constant in space and time */ 00800 00801 /******** determine new unsat potentials */ 00802 SAT_WT_HEAD[cellLoc] = SAT_WATER[cellLoc]/HP_HYD_POROSITY[HAB[cellLoc]]; 00803 UNSAT_DEPTH[cellLoc] = SED_ELEV[cellLoc]-SAT_WT_HEAD[cellLoc]; 00804 UNSAT_CAP[cellLoc] = UNSAT_DEPTH[cellLoc]*HP_HYD_POROSITY[HAB[cellLoc]]; 00805 00806 UNSAT_MOIST_PRP[cellLoc] = 00807 ( UNSAT_CAP[cellLoc]>0.0 ) ? 00808 ( Min(UNSAT_WATER[cellLoc]/UNSAT_CAP[cellLoc],1.0) ) : 00809 ( 1.0); 00810 /* determining the pathway of flow of surface water depending on depth 00811 of an unsat zone relative to the surface water */ 00812 SAT_VS_UNSAT[cellLoc] = 1/Exp(100.0*Max((SURFACE_WAT[cellLoc]-UNSAT_DEPTH[cellLoc]),0.0)); 00813 /* empirical data of a (0-1) control function, the proportion of maximum vertical water infiltration rate through soil (dependent var) as a function of soil moisture proportion (0-1) (independent var) */ 00814 UNSAT_HYD_COND_CF[cellLoc] = graph7(0x0,UNSAT_MOIST_PRP[cellLoc] ); 00815 /* field capacity = porosity - specific yield; spec yield== proportion of total soil vol 00816 that represents water that can be moved by gravity */ 00817 field_cap = (HP_HYD_POROSITY[HAB[cellLoc]]-HP_HYD_SPEC_YIELD[HAB[cellLoc]]); 00818 /* unsat_avail is proportion of water in pore space available for gravitational flow (above field capacity) */ 00819 /* e.g., when moisture prop in pore space <= field_cap/pore_space, no percolation */ 00820 /* using old moisture proportion (hasn't changed unless unsat zone was replaced by sat water) */ 00821 UNSAT_AVAIL[cellLoc] = Max(UNSAT_MOIST_PRP[cellLoc] 00822 -(field_cap)/HP_HYD_POROSITY[HAB[cellLoc]],0.0); 00823 UNSAT_WT_POT[cellLoc] = Max(UNSAT_CAP[cellLoc]-UNSAT_WATER[cellLoc],0.0); 00824 00825 /******** now determine the potential total transpiration and evaporation */ 00826 /* Potential ET is input data used in SFWMM v5.4 */ 00827 /* GP_calibET is an adjustable calibration parameter (close to 1.0, adjusted in global parameter input file) */ 00828 HYD_EVAP_CALC[cellLoc] = boundcond_ETp[cellLoc] * 0.0001*GP_calibET; /* tenths of mm *0.0001 = m */ 00829 00830 /* Leaf Area Index (LAI) of emergent macrophytes: this effective LAI estimates leaf area index that is above ponded surface water */ 00831 LAI_eff[cellLoc] = 00832 (MAC_HEIGHT[cellLoc]>0.0) ? 00833 (Max(1.0 - SURFACE_WAT[cellLoc]/MAC_HEIGHT[cellLoc], 0.0)*MAC_LAI[cellLoc]) : 00834 (0.0) ; 00835 00836 /* control function (0-1) of relative magnitude of the effective Leaf Area Index */ 00837 f_LAI_eff = exp(-LAI_eff[cellLoc]); 00838 00839 00840 HYD_TOT_POT_TRANSP[cellLoc] = HYD_EVAP_CALC[cellLoc] * (1.0-f_LAI_eff); 00841 00842 /* 0-1 control function of saturated water available to roots - capillary draw when roots don't quite reach down to water table */ 00843 SatWat_Root_CF = Exp(-10.0* Max(UNSAT_DEPTH[cellLoc]-HP_NPHBIO_ROOTDEPTH[HAB[cellLoc]],0.0) ); 00844 /* HYDrologic, control function (0-1) of proportion of WATer in upper soil profile that is AVAILable for plant uptake, including unsaturated storage withdrawal, and small capillary withdrawal from saturated storage, depending on relative depths */ 00845 HYD_WATER_AVAIL[cellLoc] = (UNSAT_DEPTH[cellLoc] > HP_NPHBIO_ROOTDEPTH[HAB[cellLoc]] ) ? 00846 ( Max(UNSAT_MOIST_PRP[cellLoc], SatWat_Root_CF) ) : 00847 ( 1.0 ); 00848 /* empirical data of a (0-1) control function, the proportion of water available to plants (dependent var) as a function of proportion (0-1) of water available upper soil profile (independent var) (generally, simply 1:1 relationship) */ 00849 MAC_WATER_AVAIL_CF[cellLoc] = graph8(0x0,HYD_WATER_AVAIL[cellLoc]); 00850 00851 /******** next calc the potential and actual flows */ 00852 /* unsatLoss(1) unsat to sat percolation */ 00853 /*unsat to sat flow here only includes percolation (rising water table accomodated in update after horiz fluxes) */ 00854 UNSAT_PERC[cellLoc] = Min(HP_HYD_RCINFILT[HAB[cellLoc]]*UNSAT_HYD_COND_CF[cellLoc],UNSAT_AVAIL[cellLoc]*UNSAT_WATER[cellLoc]); 00855 UNSAT_TO_SAT_FL[cellLoc] = 00856 ( (UNSAT_PERC[cellLoc])*DT > UNSAT_WATER[cellLoc] ) ? 00857 ( UNSAT_WATER[cellLoc]/DT ) : 00858 ( UNSAT_PERC[cellLoc]); 00859 /* unsatLoss(2) unsat to atmosph transpiration */ 00860 HYD_UNSAT_POT_TRANS[cellLoc] = (UNSAT_DEPTH[cellLoc] > HP_NPHBIO_ROOTDEPTH[HAB[cellLoc]] ) ? 00861 ( HYD_TOT_POT_TRANSP[cellLoc]*MAC_WATER_AVAIL_CF[cellLoc] ) : 00862 (0.0); /* no unsat transp if roots are in saturated zone */ 00863 UNSAT_TRANSP[cellLoc] = 00864 ( (HYD_UNSAT_POT_TRANS[cellLoc]+UNSAT_TO_SAT_FL[cellLoc])*DT>UNSAT_WATER[cellLoc] ) ? 00865 ( (UNSAT_WATER[cellLoc]-UNSAT_TO_SAT_FL[cellLoc]*DT)/DT ) : 00866 ( HYD_UNSAT_POT_TRANS[cellLoc]); 00867 00868 /* satLoss(1) sat to deep aquifer recharge **RATE parameter IS ALWAYS SET to ZERO *****/ 00869 SAT_WT_RECHG[cellLoc] = 00870 ( GP_HYD_RCRECHG*HP_HYD_SPEC_YIELD[HAB[cellLoc]]/HP_HYD_POROSITY[HAB[cellLoc]]*DT>SAT_WATER[cellLoc] ) ? 00871 ( SAT_WATER[cellLoc]/DT ) : 00872 ( GP_HYD_RCRECHG*HP_HYD_SPEC_YIELD[HAB[cellLoc]]/HP_HYD_POROSITY[HAB[cellLoc]]); 00873 00874 /* sat to surf upflow when gwater exceeds soil capacity due to lateral inflows 00875 accomodated in gwFluxes */ 00876 00877 /* satLoss(2) sat to unsat with lowering water table due to recharge loss ONLY (horiz outflows accomodated in gwFluxes) 00878 (leaves field capacity amount in unsat zone)*/ 00879 SAT_TO_UNSAT_FL[cellLoc] = 00880 ( SAT_WT_RECHG[cellLoc]*field_cap/HP_HYD_POROSITY[HAB[cellLoc]]*DT > SAT_WATER[cellLoc] ) ? 00881 ( (SAT_WATER[cellLoc])/DT ) : 00882 ( SAT_WT_RECHG[cellLoc]*field_cap/HP_HYD_POROSITY[HAB[cellLoc]]) ; 00883 /* satLoss(3) sat to atmosph */ 00884 HYD_SAT_POT_TRANS[cellLoc] = HYD_TOT_POT_TRANSP[cellLoc]*SatWat_Root_CF; 00885 SAT_WT_TRANSP[cellLoc] = 00886 ( (HYD_SAT_POT_TRANS[cellLoc]+SAT_TO_UNSAT_FL[cellLoc])*DT > SAT_WATER[cellLoc] ) ? 00887 ( (SAT_WATER[cellLoc]-SAT_TO_UNSAT_FL[cellLoc]*DT)/DT ) : 00888 ( HYD_SAT_POT_TRANS[cellLoc]); 00889 00890 /* sfwatLoss(1) sf to sat */ 00891 /* downflow to saturated assumed to occur in situations with small 00892 unsat layer overlain by significant surface water (SAT_VS_UNSAT very small), 00893 with immediate hydraulic connectivity betweent the two storages */ 00894 SF_WT_TO_SAT_DOWNFLOW[cellLoc] = 00895 ( (1.0-SAT_VS_UNSAT[cellLoc])*UNSAT_WT_POT[cellLoc]*DT>SURFACE_WAT[cellLoc] ) ? 00896 ( SURFACE_WAT[cellLoc]/DT ) : 00897 ( (1.0-SAT_VS_UNSAT[cellLoc])*UNSAT_WT_POT[cellLoc]); 00898 /* sfwatLoss(2) sf to unsat infiltration (sat_vs_unsat splits these losses to groundwater, but downflow to sat is given priority) */ 00899 SF_WT_POT_INF[cellLoc] = 00900 ( (SAT_VS_UNSAT[cellLoc]*HP_HYD_RCINFILT[HAB[cellLoc]]+SF_WT_TO_SAT_DOWNFLOW[cellLoc])*DT>SURFACE_WAT[cellLoc] ) ? 00901 ( (SURFACE_WAT[cellLoc]-SF_WT_TO_SAT_DOWNFLOW[cellLoc]*DT)/DT ) : 00902 ( SAT_VS_UNSAT[cellLoc]*HP_HYD_RCINFILT[HAB[cellLoc]]); 00903 SF_WT_INFILTRATION[cellLoc] = 00904 ( SF_WT_POT_INF[cellLoc]*DT > (UNSAT_WT_POT[cellLoc]-SF_WT_TO_SAT_DOWNFLOW[cellLoc]*DT) ) ? 00905 ((UNSAT_WT_POT[cellLoc]-SF_WT_TO_SAT_DOWNFLOW[cellLoc]*DT)/DT ) : 00906 ( SF_WT_POT_INF[cellLoc]); 00907 sfwat_pr1 = SF_WT_INFILTRATION[cellLoc]+SF_WT_TO_SAT_DOWNFLOW[cellLoc]; 00908 /* sfwatLoss(3) sf to atmosph */ 00909 SF_WT_EVAP[cellLoc] = 00910 ( (f_LAI_eff*HYD_EVAP_CALC[cellLoc]+sfwat_pr1 )*DT>SURFACE_WAT[cellLoc] ) ? 00911 ( (SURFACE_WAT[cellLoc]-sfwat_pr1*DT)/DT ) : 00912 ( f_LAI_eff*HYD_EVAP_CALC[cellLoc]); 00913 00914 00915 /******** then update the state variable storages */ 00916 00917 /* unsat loss priority: percolation, transpiration */ 00918 /* calc unsaturated storage state var (m) */ 00919 UNSAT_WATER[cellLoc] = UNSAT_WATER[cellLoc] 00920 + (SAT_TO_UNSAT_FL[cellLoc] + SF_WT_INFILTRATION[cellLoc] 00921 - UNSAT_TO_SAT_FL[cellLoc] - UNSAT_TRANSP[cellLoc]) * DT; 00922 00923 /* sat loss priority: recharge to deep aquifer, re-assign to unsat with lowered water table, transpiration */ 00924 /* calc saturated storage state var (m) */ 00925 SAT_WATER[cellLoc] = SAT_WATER[cellLoc] 00926 + (UNSAT_TO_SAT_FL[cellLoc] + SF_WT_TO_SAT_DOWNFLOW[cellLoc] 00927 - SAT_WT_TRANSP[cellLoc] - SAT_TO_UNSAT_FL[cellLoc] - SAT_WT_RECHG[cellLoc]) * DT; 00928 00929 /* sfwat loss priority: downflow to replace groundwater loss, infiltration to unsat, evaporation */ 00930 /* calc surface storage state var (m) */ 00931 SURFACE_WAT[cellLoc] = SURFACE_WAT[cellLoc] 00932 + (SF_WT_FROM_RAIN[cellLoc] 00933 - SF_WT_EVAP[cellLoc] - SF_WT_INFILTRATION[cellLoc] - SF_WT_TO_SAT_DOWNFLOW[cellLoc]) * DT; 00934 00935 /******** lastly, update of head calcs, unsat capacity, moisture proportion, etc. 00936 in order to calc water in diff storages for solute concentrations */ 00937 SAT_WT_HEAD[cellLoc] = SAT_WATER[cellLoc]/HP_HYD_POROSITY[HAB[cellLoc]]; 00938 UNSAT_DEPTH[cellLoc] = Max(SED_ELEV[cellLoc]-SAT_WT_HEAD[cellLoc],0.0); 00939 UNSAT_CAP[cellLoc] = UNSAT_DEPTH[cellLoc]*HP_HYD_POROSITY[HAB[cellLoc]]; 00940 00941 UNSAT_MOIST_PRP[cellLoc] = 00942 ( UNSAT_CAP[cellLoc]>0.0 ) ? 00943 ( Min(UNSAT_WATER[cellLoc]/UNSAT_CAP[cellLoc],1.0) ) : 00944 ( 1.0); 00945 HYD_DOM_ACTWAT_VOL[cellLoc] = ( Min(HP_DOM_MAXDEPTH[HAB[cellLoc]],UNSAT_DEPTH[cellLoc])*UNSAT_MOIST_PRP[cellLoc] + 00946 Max(HP_DOM_MAXDEPTH[HAB[cellLoc]]-UNSAT_DEPTH[cellLoc], 0.0)*HP_HYD_POROSITY[HAB[cellLoc]] ) 00947 *CELL_SIZE; 00948 /* flag for presence of small amount of water storage in the active zone must be present */ 00949 HYD_DOM_ACTWAT_PRES[cellLoc] = 00950 ( HYD_DOM_ACTWAT_VOL[cellLoc] > CELL_SIZE*GP_DetentZ ) ? 00951 ( 1.0 ) : ( 0.0); 00952 HYD_SED_WAT_VOL[cellLoc] = (SAT_WATER[cellLoc]+UNSAT_WATER[cellLoc])*CELL_SIZE; 00953 SFWT_VOL[cellLoc] = SURFACE_WAT[cellLoc]*CELL_SIZE; 00954 00955 HydTotHd[cellLoc] = SAT_WT_HEAD[cellLoc]+SURFACE_WAT[cellLoc]; /* only used for reporting purposes */ 00956 /* v2.8.2 new variable, for reporting purposes only: positive/negative water depth relative to land elevation (stage minus land elevation) */ 00957 HydRelDepPosNeg[cellLoc] = HydTotHd[cellLoc] - SED_ELEV[cellLoc]; 00958 00959 /* at the square of xx% of the macrophyte's height, the manning's n 00960 calc will indicate the macrophyte *starting* to bend over, 00961 starting to offer increasingly less resistance */ 00962 mann_height = (GP_mann_height_coef*MAC_HEIGHT[cellLoc])*(GP_mann_height_coef*MAC_HEIGHT[cellLoc]); 00963 N_density = Max(HP_MAC_MAXROUGH[HAB[cellLoc]] * MAC_REL_BIOM[cellLoc], HP_MAC_MINROUGH[HAB[cellLoc]] ); 00964 /* manning's n for overland (horiz) flow */ 00965 mann_height = Max(mann_height,0.01); /* ensure that even in absence of veg, there is miniscule (1 cm in model grid cell) height related to some form of veg */ 00966 HYD_MANNINGS_N[cellLoc] = Max(-Abs((N_density-HP_MAC_MINROUGH[HAB[cellLoc]]) 00967 *(pow(2.0,(1.0-SURFACE_WAT[cellLoc]/mann_height))-1.0) ) 00968 + N_density,HP_MAC_MINROUGH[HAB[cellLoc]]); 00969 00970 /* sum of transpiration for output only */ 00971 HYD_TRANSP[cellLoc] = UNSAT_TRANSP[cellLoc]+SAT_WT_TRANSP[cellLoc]; 00972 HYD_ET[cellLoc] = HYD_TRANSP[cellLoc]+SF_WT_EVAP[cellLoc]; 00973 00974 } 00975 } 00976 } 00977 }
| void cell_dyn8 | ( | int | step | ) |
Macrophyte (vertical) dynamics.
Temporal dynamics of carbon and phosphorus including growth, mortality, etc of macrophyte community
- Parameters:
-
step The current iteration number
Definition at line 1002 of file UnitMod.c.
References AIR_TEMP, CELL_SIZE, conv_kgTOg, conv_kgTOmg, debug, DT, dynERRORnum, Exp, GP_MAC_REFUG_MULT, GP_mac_uptake_coef, GP_MinCheck, HAB, HP_MAC_KSP, HP_MAC_LIGHTSAT, HP_MAC_MAXHT, HP_MAC_MAXLAI, HP_MAC_SALIN_THRESH, HP_MAC_TEMPOPT, HP_MAC_TRANSLOC_RC, HP_MAC_WAT_TOLER, HP_NPHBIO_IC_CTOOM, HP_NPHBIO_IC_PC, HP_NPHBIO_MAX, HP_PHBIO_IC_CTOOM, HP_PHBIO_IC_PC, HP_PHBIO_MAX, HP_PHBIO_RCMORT, HP_PHBIO_RCNPP, HYD_DOM_ACTWAT_PRES, HYD_DOM_ACTWAT_VOL, HYD_SED_WAT_VOL, MAC_HEIGHT, MAC_LAI, MAC_LIGHT_CF, MAC_MAX_BIO, MAC_NOPH_BIOMAS, mac_nph_CtoOM, mac_nph_OM, mac_nph_P, mac_nph_PC, mac_nph_PC_rep, MAC_NUT_CF, MAC_PH_BIOMAS, mac_ph_CtoOM, mac_ph_OM, mac_ph_P, mac_ph_PC, mac_ph_PC_rep, MAC_PROD_CF, MAC_REL_BIOM, MAC_SALT_CF, MAC_TEMP_CF, MAC_TOT_BIOM, MAC_WATER_AVAIL_CF, MAC_WATER_CF, Max, Min, msgStr, NPHBIO_AVAIL, NPHBIO_MORT, nphbio_mort_OM, nphbio_mort_P, NPHBIO_MORT_POT, NPHBIO_REFUGE, NPHBIO_SAT, nphbio_transl_OM, nphbio_transl_P, NPHBIO_TRANSLOC, NPHBIO_TRANSLOC_POT, ON_MAP, PHBIO_AVAIL, PHBIO_MORT, phbio_mort_OM, phbio_mort_P, PHBIO_MORT_POT, PHBIO_NPP, phbio_npp_OM, phbio_npp_P, PHBIO_REFUGE, PHBIO_SAT, phbio_transl_OM, phbio_transl_P, PHBIO_TRANSLOC, PHBIO_TRANSLOC_POT, s0, s1, SAL_SED_WT, SAL_SF_WT, salinLow, SimTime, SOLRADGRD, SURFACE_WAT, T, tempCF(), simTime::TIME, TP_SED_CONC, TP_SED_WT, TP_SED_WT_AZ, TP_SEDWT_CONCACT, TP_SEDWT_CONCACTMG, TP_SEDWT_UPTAKE, True, and WriteMsg().
Referenced by call_cell_dyn().
01003 { 01004 int ix, iy, cellLoc; 01005 float reduc, NPP_P, min_litTemp, nphbio_ddep, phbio_ddep, MAC_PHtoNPH, MAC_PHtoNPH_Init; 01006 float Sal_Mean; 01007 #define salinLow 0.1 /* v2.8.5 minimal salinity value (mg/L=ppt), below which growth isn't really affected for any macrophyre species */ 01008 01009 01010 for(ix=1; ix<=s0; ix++) { 01011 for(iy=1; iy<=s1; iy++) { 01012 01013 if(ON_MAP[cellLoc= T(ix,iy)]) { 01014 01015 /* these thresholds need updating when a habitat type of a grid cell changes */ 01016 MAC_MAX_BIO[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]]+HP_PHBIO_MAX[HAB[cellLoc]]; 01017 NPHBIO_REFUGE[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]]*GP_MAC_REFUG_MULT; 01018 NPHBIO_SAT[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]]*0.9; 01019 PHBIO_REFUGE[cellLoc] = HP_PHBIO_MAX[HAB[cellLoc]]*GP_MAC_REFUG_MULT; 01020 PHBIO_SAT[cellLoc] = HP_PHBIO_MAX[HAB[cellLoc]]*0.9; 01021 /* various control functions for production calc */ 01022 MAC_LIGHT_CF[cellLoc] = SOLRADGRD[cellLoc]/HP_MAC_LIGHTSAT[HAB[cellLoc]] 01023 *Exp(1.0-SOLRADGRD[cellLoc]/HP_MAC_LIGHTSAT[HAB[cellLoc]]); 01024 MAC_TEMP_CF[cellLoc] = tempCF(0, 0.20, HP_MAC_TEMPOPT[HAB[cellLoc]], 5.0, 40.0, AIR_TEMP[cellLoc]); 01025 HP_MAC_WAT_TOLER[HAB[cellLoc]] = Max(HP_MAC_WAT_TOLER[HAB[cellLoc]],0.005); /* water tolerance is supposed to be non-zero; set to 5mm if user input a 0 */ 01026 MAC_WATER_CF[cellLoc] = Min(MAC_WATER_AVAIL_CF[cellLoc], 01027 Max(1.0-Max( (SURFACE_WAT[cellLoc]-HP_MAC_WAT_TOLER[HAB[cellLoc]]) 01028 /HP_MAC_WAT_TOLER[HAB[cellLoc]],0.0),0.0)); 01029 MAC_NUT_CF[cellLoc] = 01030 Exp(-GP_mac_uptake_coef * Max(HP_MAC_KSP[HAB[cellLoc]]-TP_SEDWT_CONCACTMG[cellLoc], 0.0) 01031 /HP_MAC_KSP[HAB[cellLoc]]) ; /* mg/L */ 01032 /* v2.8.5, re-instituted use of salinity control on mac production */ 01033 /* NOTE the placeholder salinity CF here !!! */ 01034 Sal_Mean = (SAL_SED_WT[cellLoc]+SAL_SF_WT[cellLoc])/2.0; 01035 MAC_SALT_CF[cellLoc] = (Sal_Mean > HP_MAC_SALIN_THRESH[HAB[cellLoc]]) ? (Max(1.0-0.1*Sal_Mean/HP_MAC_SALIN_THRESH[HAB[cellLoc]],0.0) ) : (1.0); 01036 Max( 1.0-Max(Sal_Mean-(HP_MAC_SALIN_THRESH[HAB[cellLoc]]+salinLow),0.0)/(HP_MAC_SALIN_THRESH[HAB[cellLoc]]+salinLow), 0.0) ; 01037 01038 min_litTemp = Min(MAC_LIGHT_CF[cellLoc], MAC_TEMP_CF[cellLoc]); 01039 MAC_PROD_CF[cellLoc] = Min(min_litTemp,MAC_WATER_CF[cellLoc]) 01040 *MAC_NUT_CF[cellLoc] *MAC_SALT_CF[cellLoc]; 01041 /* net primary production, kg C/m2/d */ 01042 PHBIO_NPP[cellLoc] = HP_PHBIO_RCNPP[HAB[cellLoc]]*MAC_PROD_CF[cellLoc]*MAC_PH_BIOMAS[cellLoc] 01043 * Max( (1.0-MAC_TOT_BIOM[cellLoc]/MAC_MAX_BIO[cellLoc]), 0.0); 01044 /* P uptake is dependent on available P and relative to a maximum P:C ratio for the tissue (kg C/m^2/d * P:Cmax * dimless = kgP/m2/d ) */ 01045 NPP_P = PHBIO_NPP[cellLoc] * HP_PHBIO_IC_PC[HAB[cellLoc]] * Max(MAC_NUT_CF[cellLoc]*2.0,1.0) 01046 * Max(1.0-mac_ph_PC[cellLoc]/HP_PHBIO_IC_PC[HAB[cellLoc]], 0.0); 01047 /* check for available P mass that will be taken up from sed water in active zone (nutCF does not); v2.5 constrained TP_SED_WT_AZ to pos */ 01048 reduc = (NPP_P > 0.0) ? 01049 (Max(TP_SED_WT_AZ[cellLoc],0.0) / ( NPP_P*CELL_SIZE*DT) ) : /* within-plant variable stoichiometry */ 01050 (1.0); 01051 /* reduce the production proportionally if excess demand is found */ 01052 /* can have high conc, but low mass of P avail, in presence of high plant biomass and high demand */ 01053 if (reduc < 1.0) { 01054 PHBIO_NPP[cellLoc] *= reduc; 01055 NPP_P *= reduc; 01056 } 01057 01058 /* losses from photobio */ 01059 phbio_ddep = Max(1.0-Max( (PHBIO_SAT[cellLoc]-MAC_PH_BIOMAS[cellLoc]) 01060 /(PHBIO_SAT[cellLoc]-PHBIO_REFUGE[cellLoc]),0.0),0.0); 01061 PHBIO_AVAIL[cellLoc] = MAC_PH_BIOMAS[cellLoc]*phbio_ddep; 01062 MAC_PHtoNPH_Init = HP_PHBIO_MAX[HAB[cellLoc]] / HP_NPHBIO_MAX[HAB[cellLoc]] ; /*if habitat type changes */ 01063 MAC_PHtoNPH = (MAC_NOPH_BIOMAS[cellLoc]>0.0) ? ( MAC_PH_BIOMAS[cellLoc] / MAC_NOPH_BIOMAS[cellLoc]) : (MAC_PHtoNPH_Init); 01064 01065 NPHBIO_TRANSLOC_POT[cellLoc] = (MAC_PHtoNPH>MAC_PHtoNPH_Init) ? 01066 (exp(HP_MAC_TRANSLOC_RC[HAB[cellLoc]] *(MAC_PHtoNPH-MAC_PHtoNPH_Init)) - 1.0) : 01067 (0.0); 01068 NPHBIO_TRANSLOC[cellLoc] = 01069 ( (NPHBIO_TRANSLOC_POT[cellLoc])*DT >PHBIO_AVAIL[cellLoc] ) ? 01070 ( (PHBIO_AVAIL[cellLoc])/DT ) : 01071 ( NPHBIO_TRANSLOC_POT[cellLoc]); 01072 01073 PHBIO_MORT_POT[cellLoc] = HP_PHBIO_RCMORT[HAB[cellLoc]] *PHBIO_AVAIL[cellLoc] 01074 *(1.0 + (1.0-MAC_WATER_AVAIL_CF[cellLoc]) )/2.0; 01075 PHBIO_MORT[cellLoc] = 01076 ( (PHBIO_MORT_POT[cellLoc]+NPHBIO_TRANSLOC[cellLoc])*DT>PHBIO_AVAIL[cellLoc] ) ? 01077 ( (PHBIO_AVAIL[cellLoc]-NPHBIO_TRANSLOC[cellLoc]*DT)/DT ) : 01078 ( PHBIO_MORT_POT[cellLoc]); 01079 01080 01081 /* losses from non-photobio */ 01082 nphbio_ddep = Max(1.0-Max((NPHBIO_SAT[cellLoc]-MAC_NOPH_BIOMAS[cellLoc]) 01083 /(NPHBIO_SAT[cellLoc]-NPHBIO_REFUGE[cellLoc]),0.0),0.0); 01084 NPHBIO_AVAIL[cellLoc] = MAC_NOPH_BIOMAS[cellLoc]*nphbio_ddep; 01085 01086 PHBIO_TRANSLOC_POT[cellLoc] = (MAC_PHtoNPH<MAC_PHtoNPH_Init) ? 01087 (exp(HP_MAC_TRANSLOC_RC[HAB[cellLoc]] *(MAC_PHtoNPH_Init-MAC_PHtoNPH)) - 1.0) : 01088 (0.0); 01089 PHBIO_TRANSLOC[cellLoc] = 01090 ( (PHBIO_TRANSLOC_POT[cellLoc])*DT >NPHBIO_AVAIL[cellLoc] ) ? 01091 ( (NPHBIO_AVAIL[cellLoc])/DT ) : 01092 ( PHBIO_TRANSLOC_POT[cellLoc]); 01093 NPHBIO_MORT_POT[cellLoc] = NPHBIO_AVAIL[cellLoc]*HP_PHBIO_RCMORT[HAB[cellLoc]] 01094 * (1.0 + Max(1.0-MAC_PH_BIOMAS[cellLoc]/HP_PHBIO_MAX[HAB[cellLoc]],0.0) )/2.0; /* decreased mort w/ increasing photobio */ 01095 NPHBIO_MORT[cellLoc] = 01096 ( (PHBIO_TRANSLOC[cellLoc]+NPHBIO_MORT_POT[cellLoc])*DT>NPHBIO_AVAIL[cellLoc] ) ? 01097 ( (NPHBIO_AVAIL[cellLoc]-PHBIO_TRANSLOC[cellLoc]*DT)/DT ) : 01098 ( NPHBIO_MORT_POT[cellLoc]); 01099 01100 01101 /* calc nonphotosynthetic biomass state var (kgC/m2) */ 01102 MAC_NOPH_BIOMAS[cellLoc] = MAC_NOPH_BIOMAS[cellLoc] 01103 + (NPHBIO_TRANSLOC[cellLoc] - NPHBIO_MORT[cellLoc] 01104 - PHBIO_TRANSLOC[cellLoc] ) * DT; 01105 01106 /* calc nonphotosynthetic biomass state var (kgC/m2) */ 01107 MAC_PH_BIOMAS[cellLoc] = MAC_PH_BIOMAS[cellLoc] 01108 + (PHBIO_TRANSLOC[cellLoc] + PHBIO_NPP[cellLoc] 01109 - PHBIO_MORT[cellLoc] - NPHBIO_TRANSLOC[cellLoc]) * DT; 01110 01111 /* total biomass */ 01112 MAC_TOT_BIOM[cellLoc] = MAC_PH_BIOMAS[cellLoc]+MAC_NOPH_BIOMAS[cellLoc]; 01113 /* book-keeping calcs used in other modules */ 01114 MAC_REL_BIOM[cellLoc] = 01115 ( MAC_TOT_BIOM[cellLoc] > 0.0 ) ? 01116 MAC_TOT_BIOM[cellLoc]/MAC_MAX_BIO[cellLoc] : 01117 0.0001; 01118 MAC_HEIGHT[cellLoc] = pow(MAC_REL_BIOM[cellLoc],0.33)*HP_MAC_MAXHT[HAB[cellLoc]]; 01119 MAC_LAI[cellLoc] = MAC_REL_BIOM[cellLoc]*HP_MAC_MAXLAI[HAB[cellLoc]]; 01120 /* note that an "effective" LAI that accounts for water depth is calc'd in hydro module */ 01121 01122 /* now calc the P and organic matter associated with the C fluxes */ 01123 phbio_npp_P[cellLoc] = NPP_P; /* within-plant variable stoichiometry */ 01124 phbio_npp_OM[cellLoc] = PHBIO_NPP[cellLoc] / HP_PHBIO_IC_CTOOM[HAB[cellLoc]]; /* habitat-specfic stoichiometry */ 01125 01126 phbio_mort_P[cellLoc] = PHBIO_MORT[cellLoc] * mac_ph_PC[cellLoc]; 01127 phbio_mort_OM[cellLoc] = PHBIO_MORT[cellLoc] / mac_ph_CtoOM[cellLoc]; 01128 01129 phbio_transl_P[cellLoc] = PHBIO_TRANSLOC[cellLoc] * mac_nph_PC[cellLoc]; 01130 phbio_transl_OM[cellLoc] = PHBIO_TRANSLOC[cellLoc] / mac_nph_CtoOM[cellLoc]; 01131 01132 nphbio_transl_P[cellLoc] = NPHBIO_TRANSLOC[cellLoc] * mac_ph_PC[cellLoc]; 01133 nphbio_transl_OM[cellLoc] = NPHBIO_TRANSLOC[cellLoc] / mac_ph_CtoOM[cellLoc]; 01134 01135 nphbio_mort_P[cellLoc] = NPHBIO_MORT[cellLoc] * mac_nph_PC[cellLoc]; 01136 nphbio_mort_OM[cellLoc] = NPHBIO_MORT[cellLoc] / mac_nph_CtoOM[cellLoc]; 01137 01138 01139 /* state vars: now calc the P and OM associated with those C state vars */ 01140 mac_nph_P[cellLoc] = mac_nph_P[cellLoc] 01141 + (nphbio_transl_P[cellLoc] - nphbio_mort_P[cellLoc] 01142 - phbio_transl_P[cellLoc] ) * DT; 01143 mac_nph_PC[cellLoc] = ( (MAC_NOPH_BIOMAS[cellLoc] > 0.0) && (mac_nph_P[cellLoc] > 0.0)) ? 01144 (mac_nph_P[cellLoc] / MAC_NOPH_BIOMAS[cellLoc]) : /* these second mass checks not needed */ 01145 0.3 * HP_NPHBIO_IC_PC[HAB[cellLoc]]; /* default to 0.3 of max for habitat */ 01146 mac_nph_PC_rep[cellLoc] = (float)mac_nph_PC[cellLoc] * conv_kgTOmg; /* variable for output _rep-orting only */ 01147 01148 01149 mac_nph_OM[cellLoc] = mac_nph_OM[cellLoc] 01150 + (nphbio_transl_OM[cellLoc] - nphbio_mort_OM[cellLoc] 01151 - phbio_transl_OM[cellLoc] ) * DT; 01152 mac_nph_CtoOM[cellLoc] = ( (mac_nph_OM[cellLoc] > 0.0) && (MAC_NOPH_BIOMAS[cellLoc]>0.0)) ? 01153 (MAC_NOPH_BIOMAS[cellLoc] / mac_nph_OM[cellLoc]) : 01154 HP_NPHBIO_IC_CTOOM[HAB[cellLoc]]; 01155 01156 mac_ph_P[cellLoc] = mac_ph_P[cellLoc] 01157 + (phbio_transl_P[cellLoc] + phbio_npp_P[cellLoc] - phbio_mort_P[cellLoc] 01158 - nphbio_transl_P[cellLoc] ) * DT; 01159 mac_ph_PC[cellLoc] = ( (MAC_PH_BIOMAS[cellLoc] > 0.0) && (mac_ph_P[cellLoc]>0.0)) ? 01160 (mac_ph_P[cellLoc] / MAC_PH_BIOMAS[cellLoc]) : 01161 0.3 * HP_PHBIO_IC_PC[HAB[cellLoc]]; /* default to 0.3 of max for habitat */ 01162 mac_ph_PC_rep[cellLoc] = (float)mac_ph_PC[cellLoc] * conv_kgTOmg; /* variable for output _rep-orting only */ 01163 01164 mac_ph_OM[cellLoc] = mac_ph_OM[cellLoc] 01165 + (phbio_transl_OM[cellLoc] + phbio_npp_OM[cellLoc] - phbio_mort_OM[cellLoc] 01166 - nphbio_transl_OM[cellLoc] ) * DT; 01167 mac_ph_CtoOM[cellLoc] = ( (mac_ph_OM[cellLoc] > 0.0) && (MAC_PH_BIOMAS[cellLoc]>0.0)) ? 01168 (MAC_PH_BIOMAS[cellLoc] / mac_ph_OM[cellLoc]) : 01169 HP_PHBIO_IC_CTOOM[HAB[cellLoc]]; 01170 01171 if (debug > 0 && MAC_NOPH_BIOMAS[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg NPhoto C biomass (%f kg) in cell (%d,%d)!", 01172 SimTime.TIME, MAC_NOPH_BIOMAS[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 01173 if (debug > 0 && MAC_PH_BIOMAS[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg Photo C biomass (%f kg) in cell (%d,%d)!", 01174 SimTime.TIME, MAC_PH_BIOMAS[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 01175 if (debug > 0 && mac_nph_P[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg NPhoto P biomass (%f kg) in cell (%d,%d)!", 01176 SimTime.TIME, mac_nph_P[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 01177 if (debug > 0 && mac_ph_P[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg Photo P biomass (%f kg) in cell (%d,%d)!", 01178 SimTime.TIME, mac_ph_P[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 01179 01180 01181 /******** phosphorus removed from water here *************/ 01182 TP_SEDWT_UPTAKE[cellLoc] = phbio_npp_P[cellLoc]*CELL_SIZE; 01183 /* recalc P in sed water state variable (kg P) */ 01184 TP_SED_WT[cellLoc] = TP_SED_WT[cellLoc] - (TP_SEDWT_UPTAKE[cellLoc]) * DT; 01185 TP_SED_CONC[cellLoc] = (HYD_SED_WAT_VOL[cellLoc]>0.0) ? 01186 (TP_SED_WT[cellLoc] / HYD_SED_WAT_VOL[cellLoc]) : 01187 (0.0); 01188 01189 /* this is the active zone, where uptake, sorption, and mineralization take place */ 01190 TP_SED_WT_AZ[cellLoc] = TP_SED_WT_AZ[cellLoc] - (TP_SEDWT_UPTAKE[cellLoc]) * DT; 01191 TP_SEDWT_CONCACT[cellLoc] = ( HYD_DOM_ACTWAT_PRES[cellLoc] > 0.0) ? 01192 ( TP_SED_WT_AZ[cellLoc]/HYD_DOM_ACTWAT_VOL[cellLoc] ) : 01193 (TP_SED_CONC[cellLoc]); /* g/L */ 01194 TP_SEDWT_CONCACTMG[cellLoc] = TP_SEDWT_CONCACT[cellLoc]*conv_kgTOg; /* g/m3==mg/L */ 01195 01196 } 01197 } 01198 } 01199 }
| void cell_dyn9 | ( | int | step | ) |
Phosphorus (vertical) dynamics.
Temporal dynamics of phosphorus in water and sorbed to soil.
- Parameters:
-
step The current iteration number
Definition at line 1218 of file UnitMod.c.
References CELL_SIZE, conv_gTOmg, conv_kgTOg, debug, DEPOS_ORG_MAT, DIM, DT, dynERRORnum, GP_DetentZ, GP_MinCheck, GP_PO4toTP, GP_PO4toTPint, GP_settlVel, GP_TP_DIFFCOEF, GP_TP_DIFFDEPTH, GP_TP_IN_RAIN, GP_TP_K_INTER, GP_TP_K_SLOPE, GP_TPpart_thresh, GP_WQualMonitZ, HYD_DOM_ACTWAT_PRES, HYD_DOM_ACTWAT_VOL, HYD_SED_WAT_VOL, Max, msgStr, ON_MAP, s0, s1, SF_WT_FROM_RAIN, SF_WT_INFILTRATION, SF_WT_TO_SAT_DOWNFLOW, SFWT_VOL, SimTime, SURFACE_WAT, T, simTime::TIME, TP_Act_to_Tot, TP_Atm_Depos, TP_AtmosDepos, TP_DNFLOW, TP_DNFLOW_POT, TP_K, TP_SED_CONC, TP_SED_WT, TP_SED_WT_AZ, TP_SEDWT_CONCACT, TP_SEDWT_CONCACTMG, TP_settl, TP_SF_WT, TP_SFWT_CONC, TP_SFWT_CONC_MG, TP_SORB, TP_SORB_POT, TP_SORBCONC, TP_SORBCONC_rep, TP_SORBTION, TP_UPFLOW, TP_UPFLOW_POT, True, usrErr(), and WriteMsg().
Referenced by call_cell_dyn().
01219 { 01220 int ix, iy, cellLoc; 01221 float TPpartic, TPsettlRat, TP_settl_pot; 01222 double PO4Pconc, nonPO4Pconc; 01223 01224 for(ix=1; ix<=s0; ix++) { 01225 for(iy=1; iy<=s1; iy++) { 01226 01227 if(ON_MAP[cellLoc= T(ix,iy)]) { 01228 /* calc concentrations after horiz fluxes */ 01229 TP_SFWT_CONC[cellLoc] = 01230 ( SFWT_VOL[cellLoc] > 0.0 ) ? 01231 ( TP_SF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 01232 ( 0.0); /* used in P fluxes for mass balance */ 01233 /* using regression for predicting PO4 from TP */ 01234 PO4Pconc = Max( TP_SFWT_CONC[cellLoc]*GP_PO4toTP + 0.001*GP_PO4toTPint,0.0); /* g/l (note that intercept may be <0)*/ 01235 /* after spatial (horiz) fluxes, recalc the active zone P mass based on old active/total ratio */ 01236 TP_SED_CONC[cellLoc] = (HYD_SED_WAT_VOL[cellLoc]>0.0) ? 01237 (TP_SED_WT[cellLoc] / HYD_SED_WAT_VOL[cellLoc]) : 01238 (0.0); 01239 /* this is the active zone, where uptake, sorption, and mineralization take place */ 01240 TP_SED_WT_AZ[cellLoc] = TP_SED_CONC[cellLoc] * TP_Act_to_Tot[cellLoc] * HYD_DOM_ACTWAT_VOL[cellLoc]; 01241 TP_SEDWT_CONCACT[cellLoc] =(HYD_DOM_ACTWAT_PRES[cellLoc] > 0.0) ? 01242 ( TP_SED_WT_AZ[cellLoc]/HYD_DOM_ACTWAT_VOL[cellLoc] ) : 01243 ( TP_SED_CONC[cellLoc]); 01244 01245 /* inputs to surf P (kg P) */ 01246 /* v2.6, using negative-parameter switch in v2.8: 01247 - either use rainfall-based P deposition (parameter >= 0.0), 01248 or spatially varying, temporally constant, daily rate of total deposition (used if conc parameter is negative)*/ 01249 /* TODO - investigate methods/data for wet and dry deposition */ 01250 TP_Atm_Depos[cellLoc] = (GP_TP_IN_RAIN < 0.0) ? (TP_AtmosDepos[cellLoc]) : (SF_WT_FROM_RAIN[cellLoc]*CELL_SIZE*GP_TP_IN_RAIN*0.001) ; /* P deposition, kgP/day */ 01251 01252 /* calc various loss and/or vertical exchange potentials (kg P) */ 01253 /* diffusive flux */ 01254 TP_UPFLOW_POT[cellLoc] = /* advective upflow has been handled in surf-ground integration in fluxes.c */ 01255 Max((TP_SEDWT_CONCACT[cellLoc]-PO4Pconc) 01256 *GP_TP_DIFFCOEF*8.64/GP_TP_DIFFDEPTH*CELL_SIZE,0.0); /* document this diffusion constant multiplier (involving conversion from sec/day) */ 01257 TP_UPFLOW[cellLoc] = 01258 ( (TP_UPFLOW_POT[cellLoc])*DT>TP_SED_WT_AZ[cellLoc] ) ? 01259 ( (TP_SED_WT_AZ[cellLoc])/DT ) : 01260 ( TP_UPFLOW_POT[cellLoc]); 01261 /* units = mgP/L */ 01262 TP_K[cellLoc] = Max(GP_TP_K_SLOPE*TP_SORBCONC[cellLoc]+GP_TP_K_INTER,0.0); 01263 /*fix rate */ 01264 TP_SORB_POT[cellLoc] = 01265 ( HYD_DOM_ACTWAT_PRES[cellLoc]>0.0 ) ? 01266 ( (double) 0.001 01267 *(TP_K[cellLoc]*(pow(Max(TP_SEDWT_CONCACT[cellLoc],0.0),0.8) ) 01268 *0.001*(DEPOS_ORG_MAT[cellLoc]*CELL_SIZE+DIM[cellLoc])-TP_SORB[cellLoc] ) ) : 01269 ( 0.0); 01270 if (TP_SORB_POT[cellLoc]>0.0) { 01271 TP_SORBTION[cellLoc] = 01272 ( (TP_SORB_POT[cellLoc]+TP_UPFLOW[cellLoc])*DT>TP_SED_WT_AZ[cellLoc] ) ? 01273 ( (TP_SED_WT_AZ[cellLoc]-TP_UPFLOW[cellLoc]*DT)/DT ) : 01274 ( TP_SORB_POT[cellLoc]); 01275 } 01276 else { /* neg sorption, loss from sorb variable*/ 01277 TP_SORBTION[cellLoc] = 01278 ( (-TP_SORB_POT[cellLoc])*DT>TP_SORB[cellLoc] ) ? 01279 ( (-TP_SORB[cellLoc])/DT ) : 01280 ( TP_SORB_POT[cellLoc]); 01281 } 01282 01283 /* diffusive + advective flux */ 01284 TP_DNFLOW_POT[cellLoc] = (SF_WT_INFILTRATION[cellLoc]+SF_WT_TO_SAT_DOWNFLOW[cellLoc]) 01285 *CELL_SIZE*TP_SFWT_CONC[cellLoc] 01286 + Max((PO4Pconc-TP_SEDWT_CONCACT[cellLoc]) 01287 *GP_TP_DIFFCOEF*8.64/GP_TP_DIFFDEPTH*CELL_SIZE,0.0) ; 01288 TP_DNFLOW[cellLoc] = 01289 ( ( TP_DNFLOW_POT[cellLoc])*DT > TP_SF_WT[cellLoc] ) ? 01290 ( ( TP_SF_WT[cellLoc])/DT ) : 01291 ( TP_DNFLOW_POT[cellLoc]); 01292 /* calc P in sed water state variables (kg P) */ 01293 TP_SED_WT[cellLoc] = TP_SED_WT[cellLoc] + 01294 ( TP_DNFLOW[cellLoc] - TP_UPFLOW[cellLoc] - TP_SORBTION[cellLoc]) * DT; 01295 /* this is the active zone, where uptake, sorption, and mineralization take place */ 01296 TP_SED_WT_AZ[cellLoc] = TP_SED_WT_AZ[cellLoc] + 01297 (TP_DNFLOW[cellLoc] - TP_UPFLOW[cellLoc] - TP_SORBTION[cellLoc]) * DT; 01298 01299 /* calc P in surface water state variable (kg P) */ 01300 TP_SF_WT[cellLoc] = TP_SF_WT[cellLoc] + 01301 (TP_UPFLOW[cellLoc] + TP_Atm_Depos[cellLoc] 01302 - TP_DNFLOW[cellLoc]) * DT; 01303 01304 /* calc P sorbed-to-soil state variable (kg P) */ 01305 TP_SORB[cellLoc] = TP_SORB[cellLoc] + (TP_SORBTION[cellLoc]) * DT; 01306 01307 TP_SED_CONC[cellLoc] = (HYD_SED_WAT_VOL[cellLoc]>0.0) ? 01308 (TP_SED_WT[cellLoc] / HYD_SED_WAT_VOL[cellLoc]) : 01309 (0.0); /* g/L */ 01310 TP_SEDWT_CONCACT[cellLoc] = ( HYD_DOM_ACTWAT_PRES[cellLoc] > 0.0) ? 01311 ( TP_SED_WT_AZ[cellLoc]/HYD_DOM_ACTWAT_VOL[cellLoc] ) : 01312 (TP_SED_CONC[cellLoc]); /* g/L */ 01313 TP_SEDWT_CONCACTMG[cellLoc] = TP_SEDWT_CONCACT[cellLoc]*conv_kgTOg; /* g/m^3==mg/L */ 01314 01315 TP_SORBCONC[cellLoc] = ((DEPOS_ORG_MAT[cellLoc]*CELL_SIZE + DIM[cellLoc])>0.0) ? 01316 ( (double) TP_SORB[cellLoc]*conv_kgTOg / (DEPOS_ORG_MAT[cellLoc]*CELL_SIZE + DIM[cellLoc]) ) : 01317 (0.0); /* gP/kgsoil */ 01318 01319 TP_SORBCONC_rep[cellLoc] = TP_SORBCONC[cellLoc] * conv_gTOmg; /* reporting purposes only (g/kg->mg/kg)*/ 01320 01321 TP_SFWT_CONC[cellLoc] = 01322 ( SFWT_VOL[cellLoc] > 0.0 ) ? 01323 ( TP_SF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 01324 ( 0.0); /* g/L used in P fluxes for mass balance */ 01325 TP_SFWT_CONC_MG[cellLoc] = 01326 ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? 01327 (TP_SFWT_CONC[cellLoc]*conv_kgTOg) : 01328 (0.0); /* g/m^3==mg/L used for reporting and other modules to evaluate P conc when water is present */ 01329 01330 /* the following is an empirical method to calculate settling of particulate P out of the water column 01331 into the sediments. It uses the fixed ratio of PO4 to TP, but allows for a decreasing proportion of 01332 TP to be in (large) particulate form as TP concentration drops below a chosen threshold - the sum of 01333 the TP is considered to be dissolved plus large particulate plus small particulate (that cannot settle out) */ 01334 /* mass (kg) of particulate fraction of TP, available for settling to sediments */ 01335 /* using regression for predicting PO4 from TP */ 01336 PO4Pconc = Max(TP_SFWT_CONC_MG[cellLoc]*GP_PO4toTP + GP_PO4toTPint,0.0); /* mg/l (note that intercept may be <0)*/ 01337 nonPO4Pconc = Max(TP_SFWT_CONC_MG[cellLoc]-PO4Pconc,0.0); /* non-PO4 conc, mg/l */ 01338 TPpartic = nonPO4Pconc * (1.0-exp(-nonPO4Pconc/GP_TPpart_thresh)) *0.001 * SFWT_VOL[cellLoc] ; /* kg particulate P */ 01339 01340 01341 /* settling rate, 1/d */ 01342 TPsettlRat = ( SURFACE_WAT[cellLoc] > GP_DetentZ ) ? 01343 (GP_settlVel/SURFACE_WAT[cellLoc]) : 01344 0.0; 01345 01346 /* potential settling of particulate TP (kg/d) */ 01347 TP_settl_pot = TPsettlRat * TPpartic; 01348 TP_settl[cellLoc] = 01349 ( ( TP_settl_pot)*DT > TPpartic ) ? 01350 ( (TPpartic)/DT ) : 01351 ( TP_settl_pot); 01352 /* calc P in surface water state variable (kg P) */ 01353 TP_SF_WT[cellLoc] = TP_SF_WT[cellLoc] - TP_settl[cellLoc] * DT; 01354 01355 /* various book-keeping calcs used in other modules */ 01356 /* conc surf and sed wat = kgP/m3==gP/L, another var calc for mg/L */ 01357 TP_SFWT_CONC[cellLoc] = 01358 ( SFWT_VOL[cellLoc] > 0.0 ) ? 01359 ( TP_SF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 01360 ( 0.0); /* used in P fluxes for mass balance */ 01361 TP_SFWT_CONC_MG[cellLoc] = 01362 ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? 01363 (TP_SFWT_CONC[cellLoc]*conv_kgTOg) : 01364 (0.0); /* g/m3==mg/L used for reporting and other modules to evaluate P conc when water is present */ 01365 01366 if (debug > 0 && TP_SF_WT[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg TP_SF_WT (%f kg) in cell (%d,%d)!", 01367 SimTime.TIME, TP_SF_WT[cellLoc], ix,iy ); WriteMsg( msgStr,True ); usrErr( msgStr ); dynERRORnum++; } 01368 if (debug > 0 && TP_SED_WT_AZ[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg TP_SED_WT_AZ (%f kg) in cell (%d,%d)!", 01369 SimTime.TIME, TP_SED_WT_AZ[cellLoc], ix,iy ); WriteMsg( msgStr,True ); usrErr( msgStr ); dynERRORnum++; } 01370 01371 } 01372 } 01373 } 01374 }
| void cell_dyn10 | ( | int | step | ) |
Salt/tracer (vertical) dynamics.
This module can be used as a conservative tracer if salinity is not important to objectives; replicate this module if salinity and an independent, conservative tracer, are both desired.
- Parameters:
-
step The current iteration number
Definition at line 1473 of file UnitMod.c.
References CELL_SIZE, DT, GP_CL_IN_RAIN, GP_TP_DIFFCOEF, GP_TP_DIFFDEPTH, GP_WQualMonitZ, HYD_SED_WAT_VOL, Max, ON_MAP, s0, s1, SAL_SED_WT, SAL_SF_WT, SAL_SF_WT_mb, SALT_Atm_Depos, SALT_AtmosDepos, SALT_SED_TO_SF_FLOW, SALT_SED_WT, SALT_SFWAT_DOWNFL, SALT_SFWAT_DOWNFL_POT, SALT_SURF_WT, SF_WT_FROM_RAIN, SF_WT_INFILTRATION, SF_WT_TO_SAT_DOWNFLOW, SFWT_VOL, SURFACE_WAT, and T.
Referenced by call_cell_dyn().
01474 { 01475 int ix, iy, cellLoc; 01476 double SALT_SED_TO_SF_FLOW_pot; 01477 01478 01479 for(ix=1; ix<=s0; ix++) { 01480 for(iy=1; iy<=s1; iy++) { 01481 01482 if(ON_MAP[cellLoc= T(ix,iy)]) { 01483 SAL_SF_WT_mb[cellLoc] = 01484 ( SFWT_VOL[cellLoc] > 0.0 ) ? 01485 ( SALT_SURF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 01486 ( 0.0); /* used in salt fluxes for mass balance */ 01487 SAL_SED_WT[cellLoc] = 01488 ( HYD_SED_WAT_VOL[cellLoc]>0.0 ) ? 01489 ( SALT_SED_WT[cellLoc]/HYD_SED_WAT_VOL[cellLoc] ) : 01490 ( 0.0); 01491 /* v2.8: 01492 - either use rainfall-based salt (Cl) deposition (parameter >= 0.0), 01493 or spatially varying, temporally constant, daily rate of total deposition (used if conc parameter is negative)*/ 01494 /* TODO - investigate methods/data for wet and dry deposition */ 01495 SALT_Atm_Depos[cellLoc] = (GP_CL_IN_RAIN < 0.0) ? (SALT_AtmosDepos[cellLoc]) : (SF_WT_FROM_RAIN[cellLoc]*CELL_SIZE*GP_CL_IN_RAIN*0.001) ; /* chloride deposition, kgCl/day */ 01496 01497 /* diffusive + advective flux */ 01498 SALT_SFWAT_DOWNFL_POT[cellLoc] = (SF_WT_INFILTRATION[cellLoc] + SF_WT_TO_SAT_DOWNFLOW[cellLoc]) 01499 * CELL_SIZE*SAL_SF_WT_mb[cellLoc] 01500 + Max((SAL_SF_WT_mb[cellLoc]-SAL_SED_WT[cellLoc]) 01501 * GP_TP_DIFFCOEF*8.64/GP_TP_DIFFDEPTH*CELL_SIZE,0.0) ; /* TODO: get cl diffusion parm; diffusion parms same as P */ 01502 SALT_SFWAT_DOWNFL[cellLoc] = 01503 ( SALT_SFWAT_DOWNFL_POT[cellLoc]*DT>SALT_SURF_WT[cellLoc] ) ? 01504 ( SALT_SURF_WT[cellLoc]/DT ) : 01505 ( SALT_SFWAT_DOWNFL_POT[cellLoc]); 01506 01507 /* diffusive flux */ 01508 SALT_SED_TO_SF_FLOW_pot = 01509 /* advective upflow has been handled in surf-ground integration in fluxes.c */ 01510 Max((SAL_SED_WT[cellLoc]-SAL_SF_WT_mb[cellLoc]) 01511 *GP_TP_DIFFCOEF*8.64/GP_TP_DIFFDEPTH*CELL_SIZE,0.0) ; /* TODO: get cl diffusion parm; diffusion parms same as P */ 01512 SALT_SED_TO_SF_FLOW[cellLoc] = 01513 ( SALT_SED_TO_SF_FLOW_pot*DT>SALT_SED_WT[cellLoc] ) ? 01514 ( SALT_SED_WT[cellLoc]/DT ) : 01515 ( SALT_SED_TO_SF_FLOW_pot ); 01516 /* calc state vars (kg salt) */ 01517 SALT_SED_WT[cellLoc] = SALT_SED_WT[cellLoc] 01518 + (SALT_SFWAT_DOWNFL[cellLoc] - SALT_SED_TO_SF_FLOW[cellLoc]) * DT; 01519 01520 SALT_SURF_WT[cellLoc] = SALT_SURF_WT[cellLoc] 01521 + (SALT_Atm_Depos[cellLoc] + SALT_SED_TO_SF_FLOW[cellLoc] - SALT_SFWAT_DOWNFL[cellLoc] ) * DT; 01522 01523 /* book-keeping concentration calcs, (kg/m3==g/L==ppt) used in other modules */ 01524 SAL_SF_WT_mb[cellLoc] = 01525 ( SFWT_VOL[cellLoc] > 0.0 ) ? 01526 ( SALT_SURF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 01527 ( 0.0); /* used in salt fluxes for mass balance */ 01528 SAL_SF_WT[cellLoc] = 01529 ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? 01530 ( SAL_SF_WT_mb[cellLoc] ) : 01531 ( 0.0); /* used for reporting and other modules to evaluate salinity when water is present */ 01532 SAL_SED_WT[cellLoc] = 01533 ( HYD_SED_WAT_VOL[cellLoc]>0.0 ) ? 01534 ( SALT_SED_WT[cellLoc]/HYD_SED_WAT_VOL[cellLoc] ) : 01535 ( 0.0); 01536 01537 } 01538 } 01539 } 01540 01541 }
| void cell_dyn12 | ( | int | step | ) |
FLOCculent organic matter (vertical) dynamics.
Temporal dynamics of carbon and phosphorus of flocculent organic matter of (top/overlying zone of) soil.
- Parameters:
-
step The current iteration number
Definition at line 1562 of file UnitMod.c.
References C_ALG_MORT, C_ALG_MORT_P, CELL_SIZE, conv_kgTOg, conv_kgTOmg, debug, DOM_TEMP_CF, DT, dynERRORnum, Exp, FLOC, FLOC_DECOMP, FLOC_DECOMP_POT, FLOC_DECOMP_QUAL_CF, FLOC_DEPO, FLOC_DEPO_POT, FLOC_FR_ALGAE, Floc_fr_phBio, FLOC_Z, FlocP, FlocP_DECOMP, FlocP_DEPO, FlocP_FR_ALGAE, FlocP_OM, FlocP_OMrep, FlocP_PhBio, GP_ALG_C_TO_OM, GP_calibDecomp, GP_DetentZ, GP_DOM_decomp_coef, GP_DOM_DECOMP_POPT, GP_DOM_RCDECOMP, GP_Floc_BD, GP_Floc_rcSoil, GP_FlocMax, GP_MinCheck, GP_TP_P_OM, GP_WQualMonitZ, HAB, HP_DOM_AEROBTHIN, HYD_DOM_ACTWAT_PRES, HYD_DOM_ACTWAT_VOL, HYD_SED_WAT_VOL, Max, msgStr, NC_ALG_MORT, NC_ALG_MORT_P, ON_MAP, phbio_mort_OM, phbio_mort_P, s0, s1, SFWT_VOL, SimTime, soil_MOIST_CF, SURFACE_WAT, T, simTime::TIME, TP_SED_CONC, TP_SED_MINER, TP_SED_WT, TP_SED_WT_AZ, TP_SEDWT_CONCACT, TP_SEDWT_CONCACTMG, TP_settl, TP_SF_WT, TP_SFWT_CONC, TP_SFWT_CONC_MG, TP_SFWT_MINER, True, UNSAT_DEPTH, UNSAT_MOIST_PRP, and WriteMsg().
Referenced by call_cell_dyn().
01563 { 01564 int ix, iy, cellLoc; 01565 float FlocP_DECOMP_pot, FlocP_DEPO_pot, FlocP_settl, Floc_settl; 01566 01567 for(ix=1; ix<=s0; ix++) { 01568 for(iy=1; iy<=s1; iy++) { 01569 01570 if(ON_MAP[cellLoc= T(ix,iy)]) { 01571 /* inputs (kg OM / m2) */ 01572 /* all periphyton mortality goes to floc */ 01573 FLOC_FR_ALGAE[cellLoc] = (double) (C_ALG_MORT[cellLoc]+NC_ALG_MORT[cellLoc]) 01574 /GP_ALG_C_TO_OM*0.001 ; 01575 /* all photobiomass mortality goes to floc */ 01576 Floc_fr_phBio[cellLoc] = phbio_mort_OM[cellLoc]; 01577 01578 /* all settling from water column goes to floc */ 01579 FlocP_settl = TP_settl[cellLoc] / CELL_SIZE; /* kg P/m2; */ 01580 /* the particulate P settling is assumed a fixed Redfield P:OM ratio */ 01581 Floc_settl = FlocP_settl / GP_TP_P_OM; 01582 01583 01584 /* outputs (kg OM / m2) */ 01585 FLOC_DECOMP_QUAL_CF[cellLoc] = /* use the avg conc of sed and sf water here */ 01586 Exp(-GP_DOM_decomp_coef * Max(GP_DOM_DECOMP_POPT- 01587 (TP_SFWT_CONC_MG[cellLoc]+TP_SEDWT_CONCACTMG[cellLoc])/2.0, 0.0) 01588 /GP_DOM_DECOMP_POPT) ; /* mg/L */ 01589 /* decomp in surface water has higher rate than in soil, 01590 assuming this stock is of much higer substrate quality than the total soil layer */ 01591 /* GP_calibDecomp is an adjustable calib parm */ 01592 soil_MOIST_CF[cellLoc] = (UNSAT_DEPTH[cellLoc]>HP_DOM_AEROBTHIN[HAB[cellLoc]]) ? 01593 ( Max(UNSAT_MOIST_PRP[cellLoc],0.0) ) : 01594 ( 1.0 ); 01595 FLOC_DECOMP_POT[cellLoc] = GP_calibDecomp * 10.0*GP_DOM_RCDECOMP*FLOC[cellLoc] 01596 *DOM_TEMP_CF[cellLoc] *FLOC_DECOMP_QUAL_CF[cellLoc] * soil_MOIST_CF[cellLoc]; 01597 FLOC_DECOMP[cellLoc] = 01598 ( (FLOC_DECOMP_POT[cellLoc])*DT>FLOC[cellLoc] ) ? 01599 ( (FLOC[cellLoc])/DT ) : 01600 ( FLOC_DECOMP_POT[cellLoc]); 01601 01602 /* the incorporation of the floc layer into the "true" soil layer occurs 01603 at a rate dependent on the floc depth under flooded conditions, then constant rate under dry conditions */ 01604 FLOC_DEPO_POT[cellLoc] = 01605 ( SURFACE_WAT[cellLoc] > GP_DetentZ ) ? 01606 ( FLOC_Z[cellLoc]/GP_FlocMax * FLOC[cellLoc]*GP_Floc_rcSoil ) : 01607 ( FLOC[cellLoc]*GP_Floc_rcSoil); 01608 FLOC_DEPO[cellLoc] = 01609 ( (FLOC_DEPO_POT[cellLoc]+FLOC_DECOMP[cellLoc])*DT>FLOC[cellLoc] ) ? 01610 ( (FLOC[cellLoc]-FLOC_DECOMP[cellLoc]*DT)/DT ) : 01611 ( FLOC_DEPO_POT[cellLoc]); 01612 /* calc the state var (kg OM / m2) */ 01613 FLOC[cellLoc] = FLOC[cellLoc] 01614 + ( Floc_settl + Floc_fr_phBio[cellLoc] + FLOC_FR_ALGAE[cellLoc] 01615 - FLOC_DECOMP[cellLoc] - FLOC_DEPO[cellLoc] ) * DT; 01616 01617 /* the depth of floc is dependent on a fixed floc bulk density */ 01618 FLOC_Z[cellLoc] = (double) FLOC[cellLoc] / GP_Floc_BD ; 01619 01620 01621 /* Floc phosphorus (kg P / m2) */ 01622 FlocP_FR_ALGAE[cellLoc] = (double) (NC_ALG_MORT_P[cellLoc] 01623 + C_ALG_MORT_P[cellLoc])*0.001; /* kg P/m2 */ 01624 FlocP_PhBio[cellLoc] = phbio_mort_P[cellLoc] ; 01625 01626 FlocP_DECOMP_pot = FLOC_DECOMP[cellLoc] * FlocP_OM[cellLoc]; 01627 FlocP_DECOMP[cellLoc] = 01628 ( (FlocP_DECOMP_pot)*DT>FlocP[cellLoc] ) ? 01629 ( (FlocP[cellLoc])/DT ) : 01630 ( FlocP_DECOMP_pot); 01631 FlocP_DEPO_pot = FLOC_DEPO[cellLoc] * FlocP_OM[cellLoc]; 01632 FlocP_DEPO[cellLoc] = 01633 ( (FlocP_DEPO_pot+FlocP_DECOMP[cellLoc])*DT>FlocP[cellLoc] ) ? 01634 ( (FlocP[cellLoc]-FlocP_DECOMP[cellLoc]*DT)/DT ) : 01635 ( FlocP_DEPO_pot); 01636 01637 /* calc the state var (kg P / m2) */ 01638 FlocP[cellLoc] = FlocP[cellLoc] 01639 + ( FlocP_settl + FlocP_PhBio[cellLoc] + FlocP_FR_ALGAE[cellLoc] 01640 - FlocP_DECOMP[cellLoc] - FlocP_DEPO[cellLoc] ) * DT; 01641 01642 FlocP_OM[cellLoc] = ( FLOC[cellLoc]>0.0) ? (FlocP[cellLoc]/FLOC[cellLoc]) : (0.0); /* kgP/kgOM */ 01643 FlocP_OMrep[cellLoc] = (float) FlocP_OM[cellLoc] * conv_kgTOmg; /* mgP/kgOM, variable for output _rep-orting only */ 01644 01645 if (debug > 0 && FLOC[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg FLOC OM biomass (%f kg/m2) in cell (%d,%d)!", 01646 SimTime.TIME, FLOC[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 01647 if (debug > 0 && FlocP[cellLoc] < -GP_MinCheck) { sprintf(msgStr,"Day %6.1f: ERROR - neg FLOC P biomass (%f kg/m2) in cell (%d,%d)!", 01648 SimTime.TIME, FlocP[cellLoc], ix,iy ); WriteMsg( msgStr,True ); dynERRORnum++;} 01649 01650 /* now the P gain in sediment pore water with decomp - 90% goes to porewater, 10% to sfwat */ 01651 TP_SED_MINER[cellLoc] = 0.90 * FlocP_DECOMP[cellLoc] * CELL_SIZE ; 01652 /* calc P in sed water state variables (kg P) */ 01653 TP_SED_WT[cellLoc] = TP_SED_WT[cellLoc] + 01654 (TP_SED_MINER[cellLoc]) * DT; 01655 /* this is the active zone, where uptake, sorption, and mineralization take place */ 01656 TP_SED_WT_AZ[cellLoc] = TP_SED_WT_AZ[cellLoc] + 01657 (TP_SED_MINER[cellLoc]) * DT; 01658 01659 TP_SED_CONC[cellLoc] = (HYD_SED_WAT_VOL[cellLoc]>0.0) ? 01660 (TP_SED_WT[cellLoc] / HYD_SED_WAT_VOL[cellLoc]) : 01661 (0.0); 01662 TP_SEDWT_CONCACT[cellLoc] = ( HYD_DOM_ACTWAT_PRES[cellLoc] > 0.0) ? 01663 ( TP_SED_WT_AZ[cellLoc]/HYD_DOM_ACTWAT_VOL[cellLoc] ) : 01664 (TP_SED_CONC[cellLoc]); 01665 TP_SEDWT_CONCACTMG[cellLoc] = TP_SEDWT_CONCACT[cellLoc]*conv_kgTOg; /* g/m3==mg/L */ 01666 01667 01668 /* now the P gain in surface water with decomp - 90% goes to porewater, 10% to sfwat */ 01669 TP_SFWT_MINER[cellLoc] = 0.10*FlocP_DECOMP[cellLoc] * CELL_SIZE ; 01670 /* calc P in surface water state variable (kg P) */ 01671 TP_SF_WT[cellLoc] = TP_SF_WT[cellLoc] + 01672 (TP_SFWT_MINER[cellLoc]) * DT; 01673 TP_SFWT_CONC[cellLoc] = 01674 ( SFWT_VOL[cellLoc] > 0.0 ) ? 01675 ( TP_SF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 01676 ( 0.0); /* used in P fluxes for mass balance */ 01677 TP_SFWT_CONC_MG[cellLoc] = 01678 ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? 01679 (TP_SFWT_CONC[cellLoc]*conv_kgTOg) : 01680 (0.0); /* g/m3==mg/L used for reporting and other modules to evaluate P conc when water is present */ 01681 } 01682 } 01683 } 01684 }
| void cell_dyn13 | ( | int | step | ) |
Net settling of TP loss (stand-alone).
Emulates the Everglades Water Quality Model net settling rate algorithm for TP loss. The only dynamics are phosphorus input from rain and loss from settling out of water column. This should be run WITHOUT running ecological modules:
THIS SHOULD ONLY BE RUN WITH HYDROLOGY (& tracer/salt) - DO NOT RUN BIO/CHEM MODULES.
- Parameters:
-
step The current iteration number
Definition at line 1391 of file UnitMod.c.
References CELL_SIZE, conv_kgTOg, DT, GP_TP_IN_RAIN, GP_WQMthresh, GP_WQualMonitZ, ON_MAP, s0, s1, SF_WT_FROM_RAIN, SFWT_VOL, SURFACE_WAT, T, TP_Atm_Depos, TP_AtmosDepos, TP_settl, TP_settlDays, TP_SF_WT, TP_SFWT_CONC, TP_SFWT_CONC_MG, and WQMsettlVel.
Referenced by call_cell_dyn().
01392 { 01393 int ix, iy, cellLoc; 01394 float TPsettlRat, TP_settl_pot; 01395 01396 for(ix=1; ix<=s0; ix++) { 01397 for(iy=1; iy<=s1; iy++) { 01398 01399 if(ON_MAP[cellLoc= T(ix,iy)]) { 01400 /* concentration of P in surface water used in P fluxes for mass transfer (kgP/m3==gP/L) */ 01401 TP_SFWT_CONC[cellLoc] = 01402 ( SFWT_VOL[cellLoc] > 0.0 ) ? 01403 ( TP_SF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 01404 ( 0.0); 01405 /* inputs to surf P (kg P) */ 01406 /* see notes on changes, in cell_dyn9 module */ 01407 TP_Atm_Depos[cellLoc] = (GP_TP_IN_RAIN < 0.0) ? (TP_AtmosDepos[cellLoc]) : (SF_WT_FROM_RAIN[cellLoc]*CELL_SIZE*GP_TP_IN_RAIN*0.001) ; /* P deposition, kgP/day */ 01408 01409 /* TP settling rate calculation (m/d) = 0 if water depth (m) less than depth threshold parm */ 01410 if (SURFACE_WAT[cellLoc] > GP_WQMthresh ) { 01411 TPsettlRat = WQMsettlVel[cellLoc]; 01412 TP_settlDays[cellLoc]++; 01413 } 01414 else 01415 TPsettlRat = 0.0; 01416 01417 /* before we had to put in the day accumulator*/ 01418 /* TPsettlRat = ( SURFACE_WAT[cellLoc] > GP_WQMthresh ) ? */ 01419 /* (WQMsettlVel[cellLoc]) : 0.0; */ 01420 /* potential settling of particulate TP (m/d * m^2 * kg/m^3 = kg/d) */ 01421 TP_settl_pot = TPsettlRat * CELL_SIZE * TP_SFWT_CONC[cellLoc]; 01422 01423 /* like EWQM, shows mass bal error when -> TP_settl[cellLoc] = TP_settl_pot; */ 01424 01425 /* constrain settling to be no more than kg P available in water column */ 01426 TP_settl[cellLoc] = 01427 ( ( TP_settl_pot)*DT > TP_SF_WT[cellLoc] ) ? 01428 ( (TP_SF_WT[cellLoc])/DT ) : 01429 ( TP_settl_pot); 01430 /* calc P in surface water state variable (kg P) */ 01431 TP_SF_WT[cellLoc] = TP_SF_WT[cellLoc] + 01432 ( TP_Atm_Depos[cellLoc] - TP_settl[cellLoc] ) * DT; 01433 01434 /* this was in EWQM!!! (mass balance error!): if (TP_SF_WT[cellLoc]<0.0) TP_SF_WT[cellLoc]=0.0; */ 01435 01436 /* concentration of P in surface water used in P fluxes for mass transfer (kgP/m3==gP/L) */ 01437 TP_SFWT_CONC[cellLoc] = 01438 ( SFWT_VOL[cellLoc] > 0.0 ) ? 01439 ( TP_SF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : 01440 ( 0.0); 01441 /* concentration used for reporting (e.g., in maps) when water is present. 01442 Using a new (v2.8.4) depth parm also used for all 01443 concentration reporting thresholds 01444 */ 01445 TP_SFWT_CONC_MG[cellLoc] = 01446 ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? 01447 (TP_SFWT_CONC[cellLoc]*conv_kgTOg) : 01448 (0.0); /* g/m3==mg/L */ 01449 } 01450 } 01451 } 01452 }
| float tempCF | ( | int | form, | |
| float | c1, | |||
| float | topt, | |||
| float | tmin, | |||
| float | tmax, | |||
| float | tempC | |||
| ) |
Temperature control function for biology.
- Parameters:
-
form option of Lassiter or Jorgensen form of relationship c1 curvature parameter (Lassiter) topt optimum temperature (C) tmin minimum temperature (C) tmax maximum temperature (C) tempC current temperature (C)
- Returns:
- calculation of the result (dimless, 0-1)
Definition at line 1697 of file UnitMod.c.
Referenced by cell_dyn2(), cell_dyn4(), cell_dyn8(), and init_eqns().
01698 { 01699 if (form == 1) { 01700 /* Lassiter et al. 1975, where c1 = curvature parm */ 01701 return (Exp(c1*(tempC-topt)) * pow(((tmax-tempC)/(tmax-topt)), (c1*(tmax-topt))) ); 01702 } 01703 else { 01704 /* Jorgensen 1976 */ 01705 return ( Exp(-2.3 * Abs((tempC-topt)/(topt-tmin))) ); 01706 } 01707 }
| void init_static_data | ( | void | ) |
Initialize static spatial data.
Definition at line 1710 of file UnitMod.c.
References alloc_mem_stats(), basn, BCondFlow, BIRinit(), BIRoutfiles(), ESPmodeON, ON_MAP, read_map_file(), usrErr(), usrErr0(), and WQMsettlVel.
01711 { 01712 usrErr0("Acquiring static spatial data..."); /* console message */ 01713 01714 read_map_file("ModArea",ON_MAP,'c',4.0,0.0); /* defines model area, dimless unsigned char attributes, value 1 set to 4, 0=0 */ 01715 read_map_file("BoundCond",BCondFlow,'d',1.0,0.0); /* boundary condition flow restrictions, dimless integer attributes */ 01716 /* ONLY when running the EWQM settling rate version of ELM */ 01717 if (ESPmodeON) read_map_file("basinSetVel",WQMsettlVel,'f',0.0001,0.0); /* basin-based settling rates (data in tenths mm/d, converted to m/d) */ 01718 read_map_file("basins",basn,'d',1.0,0.0); /* Basins/Indicator-Region map, dimless integer attributes */ 01719 01720 alloc_mem_stats (); /* allocate memory for budget & stats arrays (need to have read the Basin/Indicator-Region map first) */ 01721 BIRinit(); /* Set up the Basin/Indicator-Region (BIR) linkages/inheritances */ 01722 BIRoutfiles(); /* Basin/Indicator-Region output files */ 01723 01724 usrErr("Done."); 01725 }
| void init_dynam_data | ( | void | ) |
Initialize dynamic spatial data.
Definition at line 1729 of file UnitMod.c.
References Bathymetry, BulkD, DOM_BD, ELEVATION, HAB, HYD_RCCONDUCT, MAC_TOT_BIOM, read_map_file(), SURFACE_WAT, TP_AtmosDepos, TP_SEDWT_CONCACT, TPtoSOIL, UNSAT_DEPTH, usrErr(), and usrErr0().
01730 { 01731 usrErr0("Acquiring dynamic spatial data..."); /* console message */ 01732 01733 read_map_file("Elevation",ELEVATION,'f',0.01,0.0); /* positive elevation relative to zero of vertical datum; input data in cm, converted to m; (prior to ELMv2.8, datum was always NGVD 1929; some apps in v2.8+ are in NAVD88; datum is independent of source code ) */ 01734 read_map_file("HAB",HAB,'c',1.0,0.0); /* habitat (veg classifications, dimless unsigned char attributes) */ 01735 read_map_file("icMacBio",MAC_TOT_BIOM,'f',0.015,0.0); /* initial total biomass of macrophytes (data in xyz, converted to kg C/m2) */ 01736 read_map_file("icSfWt",SURFACE_WAT,'f',0.01,0.0); /* initial ponded surface water (data in cm, converted to m) */ 01737 read_map_file("icUnsat",UNSAT_DEPTH,'f',0.01,0.0); /* initial depth of unsaturated zone (data in cm, converted to m) */ 01738 read_map_file("soilTP",TPtoSOIL,'f',0.000001,0.0); /* soil TP map (data in mgP/kgSoil, converted to kgP/kgSoil) */ 01739 read_map_file("soilBD",BulkD,'f',1.0,0.0); /* soil bulk dens map (data in mgSoil/cm3 == kgSoil/m3) */ 01740 read_map_file("soil_orgBD",DOM_BD,'f',1.0,0.0); /* organic soil bulk dens map (data in mgOM/cm3 == kgOM/m3) */ 01741 read_map_file("soilTPpore",TP_SEDWT_CONCACT,'f',0.000001,0.0); /* porewater TP concentration map (data in ug/L, converted to g/L == kg/m3) */ 01742 read_map_file("AtmosPdepos",TP_AtmosDepos,'f',1.0,0.0); /* rate of atmospheric deposition of total phosphorus (data in mgP/m^2/yr, convert units later with eqn inits) */ 01743 01744 /* 2 static mapps need to be here for re-initialing w/ a multiplier (sensitivity analysis) */ 01745 read_map_file("HydrCond",HYD_RCCONDUCT,'f',1.0,0.0); /* hydraulic conductivity (no conversion, data in m/d) */ 01746 read_map_file("Bathymetry",Bathymetry,'f',0.01,0.0); /* positive bathymetry (depth) relative to zero of vertical datum used in land elevation surveys (i.e., positive depth below NGVD29 or NAVD88); all positive depth data in cm, converted to m) */ 01747 01748 usrErr("Done."); /* console message */ 01749 }
| void init_eqns | ( | void | ) |
Initialization of the model equations.
This calculates initial values of state variables, and which also provides constant values of pertinent flux equations if their corresponding cell_dyn module is not executed.
Definition at line 1757 of file UnitMod.c.
References Abs, AIR_TEMP, ALG_INCID_LIGHT, ALG_LIGHT_CF, ALG_LIGHT_EXTINCT, ALG_REFUGE, ALG_SAT, ALG_TEMP_CF, ALG_WAT_CF, Arctan, Bathymetry, boundcond_depth, boundcond_ETp, boundcond_rain, BulkD, C_ALG, C_ALG_AVAIL_MORT, C_ALG_GPP, C_ALG_MORT, C_ALG_MORT_POT, C_ALG_NPP, C_ALG_NUT_CF, C_ALG_P, C_ALG_PC, C_ALG_PROD_CF, C_ALG_RESP, C_ALG_RESP_POT, CELL_SIZE, conv_gTOmg, conv_kgTOg, conv_mgTOg, conv_mgTOkg, Cos, DAYJUL, DEPOS_ORG_MAT, DIM, DOM_BD, DOM_DECOMP, DOM_DECOMP_POT, DOM_P_OM, DOM_QUALITY_CF, DOM_SED_AEROB_Z, DOM_SED_ANAEROB_Z, DOM_TEMP_CF, DOM_Z, DOP, DOP_DECOMP, DT, ELEVATION, Exp, FLOC, FLOC_DECOMP, FLOC_DECOMP_POT, FLOC_DECOMP_QUAL_CF, FLOC_DEPO, FLOC_DEPO_POT, FLOC_Z, FlocP, FMOD(), GP_ALG_IC_MULT, GP_ALG_LIGHT_SAT, GP_ALG_PC, GP_ALG_RC_MORT, GP_ALG_RC_MORT_DRY, GP_ALG_RC_PROD, GP_ALG_RC_RESP, GP_ALG_REF_MULT, GP_ALG_SHADE_FACTOR, GP_ALG_TEMP_OPT, GP_AlgComp, GP_ALTIT, GP_C_ALG_KS_P, GP_calibGWat, GP_DATUM_DISTANCE, GP_DetentZ, GP_DOM_DECOMP_POPT, GP_DOM_DECOMP_TOPT, GP_DOM_DECOMPRED, GP_DOM_RCDECOMP, GP_Floc_BD, GP_HYD_IC_SFWAT_ADD, GP_HYD_IC_UNSAT_ADD, GP_HYD_ICUNSATMOIST, GP_HYD_RCRECHG, GP_IC_BATHY_MULT, GP_IC_BulkD_MULT, GP_IC_DOM_BD_MULT, GP_IC_ELEV_MULT, GP_IC_TPtoSOIL_MULT, GP_LATDEG, GP_MAC_IC_MULT, GP_MAC_REFUG_MULT, GP_NC_ALG_KS_P, GP_PO4toTP, GP_PO4toTPint, GP_SOLOMEGA, GP_sorbToTP, GP_TP_ICSFWAT, GP_WQualMonitZ, graph8(), H2O_TEMP, HAB, HP_ALG_MAX, HP_DOM_AEROBTHIN, HP_DOM_MAXDEPTH, HP_FLOC_IC, HP_FLOC_IC_CTOOM, HP_FLOC_IC_PC, HP_HYD_POROSITY, HP_HYD_RCINFILT, HP_MAC_KSP, HP_MAC_LIGHTSAT, HP_MAC_MAXHT, HP_MAC_MAXLAI, HP_MAC_MAXROUGH, HP_MAC_MINROUGH, HP_MAC_SALIN_THRESH, HP_MAC_TEMPOPT, HP_MAC_WAT_TOLER, HP_NPHBIO_IC_CTOOM, HP_NPHBIO_IC_PC, HP_NPHBIO_MAX, HP_NPHBIO_ROOTDEPTH, HP_PHBIO_IC_CTOOM, HP_PHBIO_IC_PC, HP_PHBIO_MAX, HP_PHBIO_RCMORT, HP_PHBIO_RCNPP, HP_SALT_ICSEDWAT, HP_SALT_ICSFWAT, HP_TP_CONC_GRAD, HYD_DOM_ACTWAT_PRES, HYD_DOM_ACTWAT_VOL, HYD_MANNINGS_N, HYD_RCCONDUCT, HYD_SED_WAT_VOL, HYD_WATER_AVAIL, Inorg_Z, LAI_eff, LATRAD, MAC_HEIGHT, MAC_LAI, MAC_LIGHT_CF, MAC_MAX_BIO, MAC_NOPH_BIOMAS, mac_nph_CtoOM, mac_nph_OM, mac_nph_P, mac_nph_PC, MAC_NUT_CF, MAC_PH_BIOMAS, mac_ph_CtoOM, mac_ph_OM, mac_ph_P, mac_ph_PC, MAC_PROD_CF, MAC_REL_BIOM, MAC_SALT_CF, MAC_TEMP_CF, MAC_TOT_BIOM, MAC_WATER_AVAIL_CF, MAC_WATER_CF, Max, Min, NC_ALG, NC_ALG_AVAIL_MORT, NC_ALG_GPP, NC_ALG_MORT, NC_ALG_MORT_POT, NC_ALG_NPP, NC_ALG_NUT_CF, NC_ALG_P, NC_ALG_PC, NC_ALG_PROD_CF, NC_ALG_RESP, NC_ALG_RESP_POT, NPHBIO_AVAIL, NPHBIO_MORT, nphbio_mort_OM, nphbio_mort_P, NPHBIO_MORT_POT, NPHBIO_REFUGE, NPHBIO_SAT, nphbio_transl_OM, nphbio_transl_P, NPHBIO_TRANSLOC, NPHBIO_TRANSLOC_POT, PHBIO_AVAIL, PHBIO_MORT, phbio_mort_OM, phbio_mort_P, PHBIO_MORT_POT, PHBIO_NPP, phbio_npp_OM, phbio_npp_P, PHBIO_REFUGE, PHBIO_SAT, phbio_transl_OM, phbio_transl_P, PHBIO_TRANSLOC, PHBIO_TRANSLOC_POT, PI, s0, s1, SAL_SED_WT, SAL_SF_WT, SALT_SED_WT, SALT_SURF_WT, SAT_VS_UNSAT, SAT_WATER, SAT_WT_HEAD, SAT_WT_RECHG, satDensity, SED_ELEV, SED_INACT_Z, SF_WT_FROM_RAIN, SF_WT_INFILTRATION, SF_WT_POT_INF, SF_WT_TO_SAT_DOWNFLOW, SFWT_VOL, SimTime, Sin, soil_MOIST_CF, SOLALPHA, SOLALTCORR, SOLBETA, SOLCOSDEC, SOLDEC, SOLDEC1, SOLELEV_SINE, SOLRAD274, SOLRADATMOS, SOLRADGRD, SOLRISSET_HA, SOLRISSET_HA1, SURFACE_WAT, T, Tan, tempCF(), simTime::TIME, TP_Act_to_Tot, TP_AtmosDepos, TP_SED_CONC, TP_SED_WT, TP_SED_WT_AZ, TP_SEDWT_CONCACT, TP_SEDWT_CONCACTMG, TP_SF_WT, TP_SFWT_CONC, TP_SFWT_CONC_MG, TP_SORB, TPtoSOIL, UNSAT_CAP, UNSAT_DEPTH, UNSAT_MOIST_PRP, UNSAT_WATER, UNSAT_WT_POT, usrErr(), and usrErr0().
01758 { 01759 int ix,iy, cellLoc; 01760 float tmp; /* used in checking nutrient availability for MichMent kinetics */ 01761 float min_litTemp; /* used to determine min of temper, light cf's for alg and macs */ 01762 float I_ISat, Z_extinct, PO4Pconc, PO4P; 01763 float MAC_PHtoNPH, MAC_PHtoNPH_Init; 01764 double soil_propOrg; 01765 #define satDensity 0.9 /* assign the relative proportion (0 - 1) of a population maximum to be the saturation density for a population */ 01766 01767 usrErr0("Initializing unit model equations..."); /* console message */ 01768 01769 SimTime.TIME = 0; 01770 DAYJUL = ( FMOD(SimTime.TIME,365.0) >0.0 ) ? ( FMOD(SimTime.TIME,365.0) ) : ( 365.0); 01771 LATRAD = ((int)(GP_LATDEG)+(GP_LATDEG-(int)(GP_LATDEG))*5.0/3.0)*PI/180.0; 01772 /* AMPL = Exp(7.42+0.045*LATRAD*180.0/PI)/3600.0; */ 01773 /* DAYLENGTH = AMPL*Sin((DAYJUL-79.0)*0.01721)+12.0; */ 01774 SOLALPHA = 32.9835-64.884*(1.0-1.3614*Cos(LATRAD)); 01775 SOLDEC1 = 0.39785*Sin(4.868961+0.017203*DAYJUL +0.033446*Sin(6.224111+0.017202*DAYJUL)); 01776 SOLCOSDEC = sqrt(1.0-pow(SOLDEC1,2.0)); 01777 SOLELEV_SINE = Sin(LATRAD)*SOLDEC1+Cos(LATRAD)*SOLCOSDEC; 01778 SOLALTCORR = (1.0-Exp(-0.014*(GP_ALTIT-274.0)/(SOLELEV_SINE*274.0))); 01779 SOLBETA = 0.715-0.3183*(1.0-1.3614*Cos(LATRAD)); 01780 SOLDEC = Arctan(SOLDEC1/sqrt(1.0-pow(SOLDEC1,2.0))); 01781 SOLRISSET_HA1 = -Tan(LATRAD)*Tan(SOLDEC); 01782 SOLRISSET_HA = ( (SOLRISSET_HA1==0.0) ) ? ( PI*0.5 ) : ( ( (SOLRISSET_HA1<0.0) ) ? ( PI+Arctan(sqrt(1.0-pow(SOLRISSET_HA1,2.0))/SOLRISSET_HA1) ) : ( Arctan(sqrt(1.0-pow(SOLRISSET_HA1,2.0))/SOLRISSET_HA1))); 01783 SOLRADATMOS = 458.37*2.0*(1.0+0.033*Cos(360.0/365.0*PI/180.0*DAYJUL))* (Cos(LATRAD)*Cos(SOLDEC)*Sin(SOLRISSET_HA) + SOLRISSET_HA*180.0/(57.296*PI)*Sin(LATRAD)*Sin(SOLDEC)); 01784 01785 for(ix=0; ix<=s0+1; ix++) { 01786 for(iy=0; iy<=s1+1; iy++) { 01787 cellLoc = T(ix,iy); 01788 01789 AIR_TEMP[cellLoc] = 25.0; /* spatial time series unavailable after 1995; globally constant in v2.2+ */ 01790 /* rainfall read from sfwmm data, remapped to ELM resolution */ 01791 boundcond_rain[cellLoc] = SF_WT_FROM_RAIN[cellLoc] = boundcond_ETp[cellLoc] = boundcond_depth[cellLoc] = 0.0; 01792 01793 /* used to have cloudiness influence on GP_SOLOMEGA, now 0 */ 01794 SOLRAD274[cellLoc] = SOLRADATMOS*(SOLBETA-GP_SOLOMEGA*0.0 )-SOLALPHA; 01795 SOLRADGRD[cellLoc] = SOLRAD274[cellLoc]+((SOLRADATMOS+1.0)-SOLRAD274[cellLoc])*SOLALTCORR; 01796 H2O_TEMP[cellLoc] = AIR_TEMP[cellLoc]; 01797 01798 ALG_REFUGE[cellLoc] = HP_ALG_MAX[HAB[cellLoc]]*GP_ALG_REF_MULT; 01799 ALG_SAT[cellLoc] = HP_ALG_MAX[HAB[cellLoc]]*0.9; 01800 01801 /* v2.3: with southern everglades topo, put bathymetry back into the model */ 01802 ELEVATION[cellLoc] = ELEVATION[cellLoc] * GP_IC_ELEV_MULT; 01803 Bathymetry[cellLoc] = Bathymetry[cellLoc] * GP_IC_BATHY_MULT; 01804 SED_ELEV[cellLoc] = ELEVATION[cellLoc] - Bathymetry[cellLoc] + GP_DATUM_DISTANCE; 01805 SED_INACT_Z[cellLoc] = SED_ELEV[cellLoc]-HP_DOM_MAXDEPTH[HAB[cellLoc]]; 01806 01807 MAC_MAX_BIO[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]]+HP_PHBIO_MAX[HAB[cellLoc]]; 01808 NPHBIO_REFUGE[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]]*GP_MAC_REFUG_MULT; 01809 NPHBIO_SAT[cellLoc] = HP_NPHBIO_MAX[HAB[cellLoc]] * satDensity; 01810 PHBIO_REFUGE[cellLoc] = HP_PHBIO_MAX[HAB[cellLoc]]*GP_MAC_REFUG_MULT; 01811 PHBIO_SAT[cellLoc] = HP_PHBIO_MAX[HAB[cellLoc]] * satDensity; 01812 } 01813 } 01814 for(ix=1; ix<s0+1; ix++) { 01815 for(iy=1; iy<s1+1; iy++) { cellLoc = T(ix,iy); 01816 /*initialization of major state variables */ 01817 01818 /* hydro */ 01819 HYD_RCCONDUCT[cellLoc] = HYD_RCCONDUCT[cellLoc] * GP_calibGWat; 01820 01821 /* map */ UNSAT_DEPTH[cellLoc] = Max(UNSAT_DEPTH[cellLoc] + GP_HYD_IC_UNSAT_ADD,0.0); /* m */ 01822 SAT_WT_HEAD[cellLoc] = SED_ELEV[cellLoc]- UNSAT_DEPTH[cellLoc]; /* m */ 01823 SAT_WATER[cellLoc] = SAT_WT_HEAD[cellLoc] * HP_HYD_POROSITY[HAB[cellLoc]]; /* m */ 01824 01825 UNSAT_WATER[cellLoc] = HP_HYD_POROSITY[HAB[cellLoc]]*UNSAT_DEPTH[cellLoc] *GP_HYD_ICUNSATMOIST; /* m */ 01826 UNSAT_CAP[cellLoc] = UNSAT_DEPTH[cellLoc]*HP_HYD_POROSITY[HAB[cellLoc]]; /* m */ 01827 UNSAT_MOIST_PRP[cellLoc] = /* dimless proportion, 0-1 */ 01828 ( UNSAT_CAP[cellLoc]>0.0 ) ? 01829 ( Min(UNSAT_WATER[cellLoc]/UNSAT_CAP[cellLoc],1.0) ) : 01830 ( 1.0); 01831 01832 /* map */ SURFACE_WAT[cellLoc] = Max(SURFACE_WAT[cellLoc] + GP_HYD_IC_SFWAT_ADD, 0.0); /* m */ 01833 SFWT_VOL[cellLoc] = SURFACE_WAT[cellLoc]*CELL_SIZE; 01834 01835 HYD_DOM_ACTWAT_VOL[cellLoc] = Max(HP_DOM_MAXDEPTH[HAB[cellLoc]]-UNSAT_DEPTH[cellLoc]* 01836 (1.0-UNSAT_MOIST_PRP[cellLoc]),0.0)*HP_HYD_POROSITY[HAB[cellLoc]]*CELL_SIZE; 01837 HYD_SED_WAT_VOL[cellLoc] = (SAT_WATER[cellLoc]+UNSAT_WATER[cellLoc])*CELL_SIZE; 01838 01839 /* soil */ 01840 /* map */ DOM_BD[cellLoc] = DOM_BD[cellLoc] * GP_IC_DOM_BD_MULT; /* kg/m3 */ 01841 /* map */ BulkD[cellLoc] = BulkD[cellLoc] * GP_IC_BulkD_MULT; /* kgOM/m3 */ 01842 01843 soil_propOrg = (double) DOM_BD[cellLoc] / BulkD[cellLoc]; 01844 DIM[cellLoc] = (1.0 - soil_propOrg) * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] * CELL_SIZE; /* kg inorganic matter */ 01845 DEPOS_ORG_MAT[cellLoc] = soil_propOrg * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] ; /* kg organic matter/m2 */ 01846 // DEPOS_ORG_MAT[cellLoc] = soil_propOrg * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] ; /* kg organic matter/m2 */ 01847 /* v2.8.4 new DEPOS_ORG_MAT init */ 01848 01849 // Inorg_Z[cellLoc] = (1.0 - soil_propOrg) * HP_DOM_MAXDEPTH[HAB[cellLoc]]; /* fixed inorganic depth (m) */ 01850 // DOM_Z[cellLoc] = HP_DOM_MAXDEPTH[HAB[cellLoc]] - Inorg_Z[cellLoc]; /* m */ 01851 Inorg_Z[cellLoc] = DIM[cellLoc] / BulkD[cellLoc] * (1.0-soil_propOrg) / CELL_SIZE ; /* kgInorg / kgSoil/m3 * proprInorg / m2 == fixed inorganic depth (m) */ 01852 DOM_Z[cellLoc] = HP_DOM_MAXDEPTH[HAB[cellLoc]] - Inorg_Z[cellLoc]; /* m */ 01853 01854 // v2.8.4 DEPOS_ORG_MAT[cellLoc] = DOM_BD[cellLoc]*DOM_Z[cellLoc]; /* (mgOM/cm3 ==> kgOM/m3) * m = kgOM/m2 */ 01855 01856 DOM_SED_AEROB_Z[cellLoc] = Min(UNSAT_DEPTH[cellLoc]+HP_DOM_AEROBTHIN[HAB[cellLoc]],HP_DOM_MAXDEPTH[HAB[cellLoc]]); /* m */ 01857 DOM_SED_ANAEROB_Z[cellLoc] = HP_DOM_MAXDEPTH[HAB[cellLoc]]-DOM_SED_AEROB_Z[cellLoc]; /* m */ 01858 01859 /* map */ TPtoSOIL[cellLoc] = TPtoSOIL[cellLoc] * GP_IC_TPtoSOIL_MULT; /* kgP/kgSoil */ 01860 DOP[cellLoc] = (double) (1.0-GP_sorbToTP) * TPtoSOIL[cellLoc] * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] ; /* kgP/kg_soil * kg_soil/m3 * m == kgP/m2 */ 01861 01862 TP_SORB[cellLoc] = (double) GP_sorbToTP * TPtoSOIL[cellLoc] * BulkD[cellLoc] * HP_DOM_MAXDEPTH[HAB[cellLoc]] * CELL_SIZE; /* dimless * kgP/kg_soil * kg_soil/m3 * m * m^2 == kgP */ 01863 01864 /* floc layer of soil */ 01865 FLOC[cellLoc] = HP_FLOC_IC[HAB[cellLoc]]; /* kg OM/m2 */ 01866 FlocP[cellLoc] = FLOC[cellLoc]*HP_FLOC_IC_PC[HAB[cellLoc]]*HP_FLOC_IC_CTOOM[HAB[cellLoc]]; /* kg P /m2 */ 01867 FLOC_Z[cellLoc] = (double) FLOC[cellLoc] / GP_Floc_BD ; /* m */ 01868 01869 /* phosphorus */ 01870 TP_AtmosDepos[cellLoc] = (double)TP_AtmosDepos[cellLoc] * conv_mgTOkg * CELL_SIZE / 365.0; /* P deposition, mgP/m2/yr * kg/mg * m2 / daysPerYear = kgP/day */ 01871 /* v2.4.4 */ TP_SFWT_CONC[cellLoc] = GP_TP_ICSFWAT * conv_mgTOg; /* mg/L * g/mg => g/L */ 01872 TP_SF_WT[cellLoc] =TP_SFWT_CONC[cellLoc] * SFWT_VOL[cellLoc]; /* kg P */ 01873 TP_SFWT_CONC_MG[cellLoc] = TP_SFWT_CONC[cellLoc] * conv_gTOmg; /* mg/L */ 01874 /* using regression for predicting PO4 from TP */ 01875 PO4Pconc = Max(TP_SFWT_CONC_MG[cellLoc]*GP_PO4toTP + GP_PO4toTPint, 0.10 * TP_SFWT_CONC_MG[cellLoc]); /* mg/L */ 01876 PO4P = PO4Pconc * SFWT_VOL[cellLoc] /conv_kgTOg; /*kg P available (from conc. in mg/l) */ 01877 01878 /* v2.4.4 */ // TP_SED_CONC[cellLoc] = GP_TP_ICSEDWAT * conv_mgTOg; /* mg/L * g/mg => g/L */ 01879 // TP_SED_WT[cellLoc] = TP_SED_CONC[cellLoc] * HYD_SED_WAT_VOL[cellLoc]; /* kg P */ 01880 // /* this is the active zone, where uptake, sorption, and mineralization take place */ 01881 // TP_Act_to_Tot[cellLoc] = 1.0 / HP_TP_CONC_GRAD[HAB[cellLoc]]; /* ratio of TP conc in active zone relative to total */ 01882 // TP_SED_WT_AZ[cellLoc] = TP_SED_CONC[cellLoc] * TP_Act_to_Tot[cellLoc] * HYD_DOM_ACTWAT_VOL[cellLoc]; /* kg P */ 01883 // TP_SEDWT_CONCACT[cellLoc] =(HYD_DOM_ACTWAT_VOL[cellLoc] > 0.0) ? 01884 // ( TP_SED_WT_AZ[cellLoc]/HYD_DOM_ACTWAT_VOL[cellLoc] ) : 01885 // ( TP_SED_CONC[cellLoc]); /* g/L */ 01886 // TP_SEDWT_CONCACTMG[cellLoc] = TP_SEDWT_CONCACT[cellLoc]*conv_gTOmg; /* mg/L */ 01887 01888 /* v2.8.4 concentration in Active Zone (TP_SEDWT_CONCACT) is initialed by map (data here are g/L) */ 01889 TP_Act_to_Tot[cellLoc] = 1.0 / HP_TP_CONC_GRAD[HAB[cellLoc]]; /* ratio of TP conc in active zone relative to total */ 01890 TP_SED_CONC[cellLoc] = (TP_Act_to_Tot[cellLoc]>0.0) ? (TP_SEDWT_CONCACT[cellLoc] / TP_Act_to_Tot[cellLoc]) : (0.0); 01891 /* map */ TP_SEDWT_CONCACT[cellLoc] =(HYD_DOM_ACTWAT_VOL[cellLoc] > 0.0) ? 01892 ( TP_SEDWT_CONCACT[cellLoc] ) : 01893 ( 0.0 ); /* g/L */ 01894 TP_SEDWT_CONCACTMG[cellLoc] = TP_SEDWT_CONCACT[cellLoc]*conv_gTOmg; /* mg/L */ 01895 TP_SED_WT[cellLoc] = TP_SED_CONC[cellLoc] * HYD_SED_WAT_VOL[cellLoc]; /* kg P */ 01896 TP_SED_WT_AZ[cellLoc] = TP_SEDWT_CONCACT[cellLoc] * HYD_DOM_ACTWAT_VOL[cellLoc]; /* kg P */ 01897 01898 01899 /* salt */ 01900 SALT_SED_WT[cellLoc] = HYD_SED_WAT_VOL[cellLoc]*HP_SALT_ICSEDWAT[HAB[cellLoc]]; 01901 SAL_SED_WT[cellLoc] = ( HYD_SED_WAT_VOL[cellLoc]>0.0 ) ? ( SALT_SED_WT[cellLoc]/HYD_SED_WAT_VOL[cellLoc] ) : ( 0.0); 01902 SALT_SURF_WT[cellLoc] = SFWT_VOL[cellLoc]*HP_SALT_ICSFWAT[HAB[cellLoc]]; 01903 SAL_SF_WT[cellLoc] = ( SURFACE_WAT[cellLoc] > GP_WQualMonitZ ) ? ( SALT_SURF_WT[cellLoc]/SFWT_VOL[cellLoc] ) : ( 0.0); 01904 01905 /* periphyton */ 01906 /* 2.4.4 */ NC_ALG[cellLoc] = HP_ALG_MAX[HAB[cellLoc]] * GP_ALG_IC_MULT * GP_ALG_REF_MULT ; /* start w/ low, refuge-level of non-calcareous (eutrophic) periphyton, g C/m2 */ 01907 /* 2.4.4 */ C_ALG[cellLoc] = HP_ALG_MAX[HAB[cellLoc]] * GP_ALG_IC_MULT * (1.0 - GP_ALG_REF_MULT); /* g C/m2 */ 01908 /* ic PC of periph in oligotrophic area is 3% of max P:C, varies across space via (0.1->1.0) map */ 01909 /* 2.4.4 */ NC_ALG_PC[cellLoc] = GP_ALG_PC; /* gP/ gC */ 01910 /* 2.4.4 */ C_ALG_PC[cellLoc] = GP_ALG_PC; /* gP/ gC */ 01911 01912 NC_ALG_P[cellLoc] = NC_ALG[cellLoc]*NC_ALG_PC[cellLoc]; /* g P/m2 */ 01913 C_ALG_P[cellLoc] = C_ALG[cellLoc]*C_ALG_PC[cellLoc]; /* g P/m2 */ 01914 01915 /* macrophytes */ 01916 /* 2.4.4 */ MAC_PH_BIOMAS[cellLoc] = MAC_TOT_BIOM[cellLoc] * GP_MAC_IC_MULT * HP_PHBIO_MAX[HAB[cellLoc]]/MAC_MAX_BIO[cellLoc]; /* kg C/m2 */ 01917 /* now calc the P and OM associated with that C */ 01918 /* 2.4.4 */ mac_ph_PC[cellLoc] = HP_PHBIO_IC_PC[HAB[cellLoc]]; 01919 mac_ph_P[cellLoc] = MAC_PH_BIOMAS[cellLoc] * mac_ph_PC[cellLoc]; /* kg P/m2 */ 01920 mac_ph_OM[cellLoc] = MAC_PH_BIOMAS[cellLoc] / HP_PHBIO_IC_CTOOM[HAB[cellLoc]]; 01921 mac_ph_CtoOM[cellLoc] = HP_PHBIO_IC_CTOOM[HAB[cellLoc]]; 01922 PHBIO_AVAIL[cellLoc] = MAC_PH_BIOMAS[cellLoc]*Max(1.0-Max((PHBIO_SAT[cellLoc]-MAC_PH_BIOMAS[cellLoc]) /(PHBIO_SAT[cellLoc]-PHBIO_REFUGE[cellLoc]),0.0),0.0); 01923 01924 /* 2.4.4 */ MAC_NOPH_BIOMAS[cellLoc] = MAC_TOT_BIOM[cellLoc] * GP_MAC_IC_MULT * HP_NPHBIO_MAX[HAB[cellLoc]]/MAC_MAX_BIO[cellLoc]; /* kg C/m2 */ 01925 /* now calc the P and OM associated with that C */ 01926 /* 2.4.4 */ mac_nph_PC[cellLoc] = HP_NPHBIO_IC_PC[HAB[cellLoc]]; 01927 mac_nph_P[cellLoc] = MAC_NOPH_BIOMAS[cellLoc] * mac_nph_PC[cellLoc]; /* kg P/m2 */ 01928 mac_nph_OM[cellLoc] = MAC_NOPH_BIOMAS[cellLoc] / HP_NPHBIO_IC_CTOOM[HAB[cellLoc]]; 01929 mac_nph_CtoOM[cellLoc] = HP_NPHBIO_IC_CTOOM[HAB[cellLoc]]; 01930 NPHBIO_AVAIL[cellLoc] = MAC_NOPH_BIOMAS[cellLoc]*Max(1.0-Max((NPHBIO_SAT[cellLoc]-MAC_NOPH_BIOMAS[cellLoc])/(NPHBIO_SAT[cellLoc]-NPHBIO_REFUGE[cellLoc]),0.0),0.0); 01931 01932 MAC_REL_BIOM[cellLoc] = ( MAC_TOT_BIOM[cellLoc] > 0 ) ? MAC_TOT_BIOM[cellLoc]/MAC_MAX_BIO[cellLoc] : 0.0; 01933 MAC_LAI[cellLoc] = MAC_REL_BIOM[cellLoc]*HP_MAC_MAXLAI[HAB[cellLoc]]; 01934 MAC_HEIGHT[cellLoc] = MAC_REL_BIOM[cellLoc]*HP_MAC_MAXHT[HAB[cellLoc]]; 01935 LAI_eff[cellLoc] = (MAC_HEIGHT[cellLoc]>0.0) ? (Max(1.0 - SURFACE_WAT[cellLoc]/MAC_HEIGHT[cellLoc], 0.0)*MAC_LAI[cellLoc]) : (0.0) ; 01936 01937 /* end of initialization of major state variables */ 01938 01939 /* *************************** */ 01940 /* These are the multiple calculations used if particular modules are turned off. \n 01941 NOTE: THIS section (of init_eqns() ) is not fully updated for properly 01942 turning off individual **interacting** *biological/chemical* modules. 01943 If one *biological/chemical* module is turned off, 01944 they all need to be turned off. (Note that cell_dyn's 3,5,6 should always be off). \n 01945 01946 *** \n 01947 The following *biological/chemical* modules must be ON or OFF as a group: 01948 (cell_dyn2 + cell_dyn4 + cell_dyn8 + cell_dyn9 + cell_dyn12) 01949 cell_dyn13, the net settling rate module, can be turned on only when above module group is off 01950 *** \n 01951 01952 In particular, the budget will 01953 not properly reflect actual dynamics if those 01954 modules are not treated as a group. 01955 */ 01956 NC_ALG_MORT_POT[cellLoc] = ( UNSAT_DEPTH[cellLoc]>0.05 ) ? ( NC_ALG[cellLoc]*GP_ALG_RC_MORT_DRY ) : ( NC_ALG[cellLoc]*GP_ALG_RC_MORT); 01957 /* calcareous periphyton */ 01958 C_ALG_MORT_POT[cellLoc] = ( UNSAT_DEPTH[cellLoc]>0.05 ) ? ( C_ALG[cellLoc]*GP_ALG_RC_MORT_DRY ) : ( C_ALG[cellLoc]*GP_ALG_RC_MORT); 01959 ALG_TEMP_CF[cellLoc] = tempCF(0, 0.20, GP_ALG_TEMP_OPT, 5.0, 40.0, H2O_TEMP[cellLoc]); 01960 NC_ALG_RESP_POT[cellLoc] = 01961 ( UNSAT_DEPTH[cellLoc]<0.05 ) ? 01962 ( GP_ALG_RC_RESP*ALG_TEMP_CF[cellLoc]*NC_ALG[cellLoc] ) : 01963 ( 0.0); 01964 NC_ALG_RESP[cellLoc] = 01965 ( NC_ALG_RESP_POT[cellLoc]*DT>NC_ALG[cellLoc] ) ? 01966 ( NC_ALG[cellLoc]/DT ) : 01967 ( NC_ALG_RESP_POT[cellLoc]); 01968 /* calcareous periphyton */ 01969 C_ALG_RESP_POT[cellLoc] = 01970 ( UNSAT_DEPTH[cellLoc]<0.05 ) ? 01971 ( GP_ALG_RC_RESP*ALG_TEMP_CF[cellLoc]*C_ALG[cellLoc] ) : 01972 ( 0.0); 01973 C_ALG_RESP[cellLoc] = 01974 ( C_ALG_RESP_POT[cellLoc]*DT>C_ALG[cellLoc] ) ? 01975 ( C_ALG[cellLoc]/DT ) : 01976 ( C_ALG_RESP_POT[cellLoc]); 01977 01978 NC_ALG_AVAIL_MORT[cellLoc] = NC_ALG[cellLoc]-ALG_REFUGE[cellLoc]; 01979 NC_ALG_MORT[cellLoc] = ( (NC_ALG_MORT_POT[cellLoc])*DT>NC_ALG_AVAIL_MORT[cellLoc] ) ? ( (NC_ALG_AVAIL_MORT[cellLoc])/DT ) : ( NC_ALG_MORT_POT[cellLoc]); 01980 /* calcareous periphyton */ 01981 C_ALG_AVAIL_MORT[cellLoc] = C_ALG[cellLoc]-ALG_REFUGE[cellLoc]; 01982 C_ALG_MORT[cellLoc] = ( (C_ALG_MORT_POT[cellLoc])*DT>C_ALG_AVAIL_MORT[cellLoc] ) ? ( (C_ALG_AVAIL_MORT[cellLoc])/DT ) : ( C_ALG_MORT_POT[cellLoc]); 01983 01984 /* light, water, temperature controls apply to calc and non-calc periphyton */ 01985 ALG_LIGHT_EXTINCT[cellLoc] = 0.01; /* light extinction coef */ 01986 /* algal self-shading implicit in density-dependent constraint function later */ 01987 ALG_INCID_LIGHT[cellLoc] = SOLRADGRD[cellLoc]*Exp(-MAC_LAI[cellLoc]*GP_ALG_SHADE_FACTOR); 01988 Z_extinct = SURFACE_WAT[cellLoc]*ALG_LIGHT_EXTINCT[cellLoc]; 01989 I_ISat = ALG_INCID_LIGHT[cellLoc]/GP_ALG_LIGHT_SAT; 01990 /* averaged over whole water column (based on Steele '65) */ 01991 ALG_LIGHT_CF[cellLoc] = ( Z_extinct > 0.0 ) ? 01992 ( 2.718/Z_extinct * (Exp(-I_ISat * Exp(-Z_extinct)) - Exp(-I_ISat)) ) : 01993 (I_ISat*Exp(1.0-I_ISat)); 01994 /* low-water growth constraint ready for something better based on data */ 01995 ALG_WAT_CF[cellLoc] = ( SURFACE_WAT[cellLoc]>0.0 ) ? ( 1.0 ) : ( 0.0); 01996 /* the 2 communities have same growth response to avail phosphorus - avail P is roughly calc'd from TP */ 01997 NC_ALG_NUT_CF[cellLoc] = PO4Pconc/(PO4Pconc+GP_NC_ALG_KS_P) ; 01998 C_ALG_NUT_CF[cellLoc] = PO4Pconc/(PO4Pconc+GP_C_ALG_KS_P); 01999 min_litTemp = Min(ALG_LIGHT_CF[cellLoc],ALG_TEMP_CF[cellLoc]); 02000 NC_ALG_PROD_CF[cellLoc] = Min(min_litTemp,ALG_WAT_CF[cellLoc])*NC_ALG_NUT_CF[cellLoc]; 02001 C_ALG_PROD_CF[cellLoc] = Min(min_litTemp,ALG_WAT_CF[cellLoc])*C_ALG_NUT_CF[cellLoc]; 02002 /* gross production of the 2 communities (gC/m2, NOT kgC/m2) */ 02003 /* density constraint contains both noncalc and calc, competition effect accentuated by calc algae */ 02004 /* used to increase calc growth by factor of 10 */ 02005 NC_ALG_GPP[cellLoc] = NC_ALG_PROD_CF[cellLoc]*GP_ALG_RC_PROD*NC_ALG[cellLoc] 02006 *Max( (1.0-(GP_AlgComp*C_ALG[cellLoc]+NC_ALG[cellLoc])/HP_ALG_MAX[HAB[cellLoc]]),0.0); 02007 C_ALG_GPP[cellLoc] = C_ALG_PROD_CF[cellLoc]*GP_ALG_RC_PROD*C_ALG[cellLoc] 02008 *Max( (1.0-( C_ALG[cellLoc]+NC_ALG[cellLoc])/HP_ALG_MAX[HAB[cellLoc]]),0.0); 02009 /* check for available P mass (the MichMent kinetics nutCF do not) */ 02010 tmp = ( (NC_ALG_GPP[cellLoc]+C_ALG_GPP[cellLoc]) > 0) ? 02011 PO4P / ( (NC_ALG_GPP[cellLoc]+C_ALG_GPP[cellLoc]) 02012 * 0.001*GP_ALG_PC*CELL_SIZE*DT) : 02013 1.0; 02014 if (tmp < 1.0) { 02015 NC_ALG_GPP[cellLoc] *= tmp; 02016 C_ALG_GPP[cellLoc] *= tmp; 02017 /* can have high conc, but low mass of P avail, in presence of high peri biomass and high demand */ 02018 /* reduce the production proportionally if excess demand is found */ 02019 } 02020 /* total of calc and noncalc algae available and their total NPP */ 02021 NC_ALG_NPP[cellLoc] = NC_ALG_GPP[cellLoc]-NC_ALG_RESP[cellLoc]; 02022 C_ALG_NPP[cellLoc] = C_ALG_GPP[cellLoc]-C_ALG_RESP[cellLoc]; 02023 02024 02025 DOM_QUALITY_CF[cellLoc] = (Max(TP_SFWT_CONC_MG[cellLoc],TP_SEDWT_CONCACTMG[cellLoc])) 02026 /GP_DOM_DECOMP_POPT; /* mg/L */ 02027 DOM_TEMP_CF[cellLoc] = Exp(0.20*(H2O_TEMP[cellLoc]-GP_DOM_DECOMP_TOPT))*pow(((40.0-H2O_TEMP[cellLoc])/(40.0-GP_DOM_DECOMP_TOPT)),(0.20*(40.0-GP_DOM_DECOMP_TOPT))); 02028 soil_MOIST_CF[cellLoc] = pow(Max(UNSAT_MOIST_PRP[cellLoc],0.0),0.75); 02029 DOM_DECOMP_POT[cellLoc] = GP_DOM_RCDECOMP*DOM_QUALITY_CF[cellLoc]*DOM_TEMP_CF[cellLoc]*DEPOS_ORG_MAT[cellLoc]*(Min(DOM_SED_AEROB_Z[cellLoc]/HP_DOM_MAXDEPTH[HAB[cellLoc]],1.0)*soil_MOIST_CF[cellLoc]+GP_DOM_DECOMPRED*Min(DOM_SED_ANAEROB_Z[cellLoc]/HP_DOM_MAXDEPTH[HAB[cellLoc]],1.0)); 02030 DOM_DECOMP[cellLoc] = ( (DOM_DECOMP_POT[cellLoc])*DT>DEPOS_ORG_MAT[cellLoc] ) ? ( (DEPOS_ORG_MAT[cellLoc])/DT ) : ( DOM_DECOMP_POT[cellLoc]); 02031 /* added for P DOM stoich */ 02032 DOP_DECOMP[cellLoc] = DOM_DECOMP[cellLoc] * DOM_P_OM[cellLoc]; 02033 02034 SAT_VS_UNSAT[cellLoc] = 1/Exp(100.0*Max((SURFACE_WAT[cellLoc]-UNSAT_DEPTH[cellLoc]),0.0)); 02035 UNSAT_WT_POT[cellLoc] = Max(UNSAT_CAP[cellLoc]-UNSAT_WATER[cellLoc],0.0); 02036 SF_WT_TO_SAT_DOWNFLOW[cellLoc] = ( (1.0-SAT_VS_UNSAT[cellLoc])*UNSAT_WT_POT[cellLoc]*DT>SURFACE_WAT[cellLoc] ) ? ( SURFACE_WAT[cellLoc]/DT ) : ( (1.0-SAT_VS_UNSAT[cellLoc])*UNSAT_WT_POT[cellLoc]); 02037 SAT_WT_RECHG[cellLoc] = ( GP_HYD_RCRECHG*DT>SAT_WATER[cellLoc] ) ? ( SAT_WATER[cellLoc]/DT ) : ( GP_HYD_RCRECHG); 02038 SF_WT_POT_INF[cellLoc] = ( (SURFACE_WAT[cellLoc]<HP_HYD_RCINFILT[HAB[cellLoc]]*DT) ) ? ( SURFACE_WAT[cellLoc]/DT ) : ( HP_HYD_RCINFILT[HAB[cellLoc]]); 02039 SF_WT_INFILTRATION[cellLoc] = ( SF_WT_POT_INF[cellLoc]*SAT_VS_UNSAT[cellLoc]*DT>UNSAT_WT_POT[cellLoc] ) ? ( (UNSAT_WT_POT[cellLoc])/DT ) : ( SF_WT_POT_INF[cellLoc]*SAT_VS_UNSAT[cellLoc]); 02040 HYD_DOM_ACTWAT_PRES[cellLoc] = ( HYD_DOM_ACTWAT_VOL[cellLoc] > 0.01 ) ? ( 1.0 ) : ( 0.0); 02041 HYD_WATER_AVAIL[cellLoc] = Min(1.0, UNSAT_MOIST_PRP[cellLoc]+Exp(-10.0*Max(UNSAT_DEPTH[cellLoc]-HP_NPHBIO_ROOTDEPTH[HAB[cellLoc]],0.0))); 02042 02043 MAC_LIGHT_CF[cellLoc] = SOLRADGRD[cellLoc]/HP_MAC_LIGHTSAT[HAB[cellLoc]]*Exp(1.0-SOLRADGRD[cellLoc]/HP_MAC_LIGHTSAT[HAB[cellLoc]]); 02044 MAC_TEMP_CF[cellLoc] = Exp(0.20*(AIR_TEMP[cellLoc]-HP_MAC_TEMPOPT[HAB[cellLoc]])) *pow(((40.0-AIR_TEMP[cellLoc])/(40.0-HP_MAC_TEMPOPT[HAB[cellLoc]])),(0.20*(40.0-HP_MAC_TEMPOPT[HAB[cellLoc]]))); 02045 MAC_WATER_AVAIL_CF[cellLoc] = graph8(0x0,HYD_WATER_AVAIL[cellLoc]); 02046 MAC_WATER_CF[cellLoc] = Min(MAC_WATER_AVAIL_CF[cellLoc], Max(1.0-Max((SURFACE_WAT[cellLoc]-HP_MAC_WAT_TOLER[HAB[cellLoc]])/HP_MAC_WAT_TOLER[HAB[cellLoc]],0.0),0.0)); 02047 MAC_NUT_CF[cellLoc] = TP_SEDWT_CONCACT[cellLoc]/(TP_SEDWT_CONCACT[cellLoc]+HP_MAC_KSP[HAB[cellLoc]]*0.001) ; 02048 02049 MAC_SALT_CF[cellLoc] = ( HP_MAC_SALIN_THRESH[HAB[cellLoc]]>0.0 ) ? ( Max( 1.0-Max(SAL_SED_WT[cellLoc]-HP_MAC_SALIN_THRESH[HAB[cellLoc]],0.0)/HP_MAC_SALIN_THRESH[HAB[cellLoc]],0.0) ) : ( Max(1.0-SAL_SED_WT[cellLoc],0.0)); 02050 min_litTemp = Min(MAC_LIGHT_CF[cellLoc], MAC_TEMP_CF[cellLoc]); 02051 MAC_PROD_CF[cellLoc] = Min(min_litTemp,MAC_WATER_CF[cellLoc]) 02052 *MAC_NUT_CF[cellLoc]*MAC_SALT_CF[cellLoc]; 02053 PHBIO_NPP[cellLoc] = HP_PHBIO_RCNPP[HAB[cellLoc]]*MAC_PROD_CF[cellLoc]*MAC_PH_BIOMAS[cellLoc]* Max( (1.0-MAC_TOT_BIOM[cellLoc]/MAC_MAX_BIO[cellLoc]), 0.0); 02054 /* check for available P mass that will be taken up (MichMent kinetics in nutCF does not) */ 02055 tmp = (PHBIO_NPP[cellLoc] > 0) ? 02056 TP_SED_WT[cellLoc] / ( PHBIO_NPP[cellLoc] * HP_NPHBIO_IC_PC[HAB[cellLoc]]*CELL_SIZE*DT) : 02057 1.0; 02058 if (tmp < 1.0) PHBIO_NPP[cellLoc] *= tmp; 02059 /* reduce the production proportionally if excess demand is found */ 02060 /* can have high conc, but low mass of P avail, in presence of high plant biomass and high demand */ 02061 /* now add the P and OM associated with that C */ 02062 phbio_npp_P[cellLoc] = PHBIO_NPP[cellLoc] * HP_PHBIO_IC_PC[HAB[cellLoc]]; /* habitat-specfic stoichiometry */ 02063 phbio_npp_OM[cellLoc] = PHBIO_NPP[cellLoc] / HP_PHBIO_IC_CTOOM[HAB[cellLoc]]; /* habitat-specfic stoichiometry */ 02064 02065 /* init ("target") ph/nph ratio and new transloc algorithm */ 02066 MAC_PHtoNPH_Init = HP_PHBIO_MAX[HAB[cellLoc]] / HP_NPHBIO_MAX[HAB[cellLoc]] ; 02067 MAC_PHtoNPH = MAC_PH_BIOMAS[cellLoc] / MAC_NOPH_BIOMAS[cellLoc]; 02068 02069 PHBIO_TRANSLOC_POT[cellLoc] = 0.0; /* (MAC_PHtoNPH<MAC_PHtoNPH_Init) ? (exp(HP_MAC_TRANSLOC_RC[HAB[cellLoc]] *(MAC_PHtoNPH_Init-MAC_PHtoNPH)) - 1.0) : (0.0) */ 02070 02071 PHBIO_TRANSLOC[cellLoc] = ( (PHBIO_TRANSLOC_POT[cellLoc])*DT >NPHBIO_AVAIL[cellLoc] ) ? ( (NPHBIO_AVAIL[cellLoc])/DT ) : ( PHBIO_TRANSLOC_POT[cellLoc]); 02072 /* now remove the P and OM associated with that C */ 02073 phbio_transl_P[cellLoc] = PHBIO_TRANSLOC[cellLoc] * mac_nph_PC[cellLoc]; 02074 phbio_transl_OM[cellLoc] = PHBIO_TRANSLOC[cellLoc] / mac_nph_CtoOM[cellLoc]; 02075 NPHBIO_MORT_POT[cellLoc] = NPHBIO_AVAIL[cellLoc]*HP_PHBIO_RCMORT[HAB[cellLoc]]*(1.0-MAC_PH_BIOMAS[cellLoc]/HP_PHBIO_MAX[HAB[cellLoc]]); 02076 NPHBIO_MORT[cellLoc] = ( (PHBIO_TRANSLOC[cellLoc]+NPHBIO_MORT_POT[cellLoc])*DT>NPHBIO_AVAIL[cellLoc] ) ? ( (NPHBIO_AVAIL[cellLoc]-PHBIO_TRANSLOC[cellLoc]*DT)/DT ) : ( NPHBIO_MORT_POT[cellLoc]); 02077 PHBIO_MORT_POT[cellLoc] = HP_PHBIO_RCMORT[HAB[cellLoc]] *PHBIO_AVAIL[cellLoc] *(1.0-MAC_WATER_AVAIL_CF[cellLoc]); 02078 /* now remove the P and OM associated with that C */ 02079 nphbio_mort_P[cellLoc] = NPHBIO_MORT[cellLoc] * mac_nph_PC[cellLoc]; 02080 nphbio_mort_OM[cellLoc] = NPHBIO_MORT[cellLoc] / mac_nph_CtoOM[cellLoc]; 02081 02082 NPHBIO_TRANSLOC_POT[cellLoc] = 0.0; /* (MAC_PHtoNPH>MAC_PHtoNPH_Init) ? (exp(HP_MAC_TRANSLOC_RC[HAB[cellLoc]] *(MAC_PHtoNPH-MAC_PHtoNPH_Init)) - 1.0) : (0.0) */ 02083 NPHBIO_TRANSLOC[cellLoc] = ( (NPHBIO_TRANSLOC_POT[cellLoc])*DT >MAC_PH_BIOMAS[cellLoc] ) ? ( (MAC_PH_BIOMAS[cellLoc])/DT ) : ( NPHBIO_TRANSLOC_POT[cellLoc]); 02084 /* now remove the P and OM associated with that C */ 02085 nphbio_transl_P[cellLoc] = NPHBIO_TRANSLOC[cellLoc] * mac_ph_PC[cellLoc]; 02086 nphbio_transl_OM[cellLoc] = NPHBIO_TRANSLOC[cellLoc] / mac_ph_CtoOM[cellLoc]; 02087 PHBIO_MORT[cellLoc] = ( (PHBIO_MORT_POT[cellLoc]+NPHBIO_TRANSLOC[cellLoc])*DT>PHBIO_AVAIL[cellLoc] ) ? ( (PHBIO_AVAIL[cellLoc]-NPHBIO_TRANSLOC[cellLoc]*DT)/DT ) : ( PHBIO_MORT_POT[cellLoc]); 02088 /* now remove the P associated with that C */ 02089 phbio_mort_P[cellLoc] = PHBIO_MORT[cellLoc] * mac_ph_PC[cellLoc]; 02090 phbio_mort_OM[cellLoc] = PHBIO_MORT[cellLoc] / mac_ph_CtoOM[cellLoc]; 02091 02092 02093 FLOC_DECOMP_QUAL_CF[cellLoc] = Min(TP_SFWT_CONC_MG[cellLoc]/GP_DOM_DECOMP_POPT,1.0) ; 02094 FLOC_DECOMP_POT[cellLoc] = GP_DOM_RCDECOMP*FLOC[cellLoc]*DOM_TEMP_CF[cellLoc] *FLOC_DECOMP_QUAL_CF[cellLoc]; 02095 FLOC_DECOMP[cellLoc] = ( (FLOC_DECOMP_POT[cellLoc])*DT>FLOC[cellLoc] ) ? ( (FLOC[cellLoc])/DT ) : ( FLOC_DECOMP_POT[cellLoc]); 02096 FLOC_DEPO_POT[cellLoc] = ( SURFACE_WAT[cellLoc] > GP_DetentZ ) ? ( FLOC[cellLoc]*0.05 ) : ( FLOC[cellLoc]/DT); 02097 FLOC_DEPO[cellLoc] = ( (FLOC_DEPO_POT[cellLoc]+FLOC_DECOMP[cellLoc])*DT>FLOC[cellLoc] ) ? ( (FLOC[cellLoc]-FLOC_DECOMP[cellLoc]*DT)/DT ) : ( FLOC_DEPO_POT[cellLoc]); 02098 02099 HYD_MANNINGS_N[cellLoc] = Max(-Abs((HP_MAC_MAXROUGH[HAB[cellLoc]]-HP_MAC_MINROUGH[HAB[cellLoc]])* (pow(2.0,(1.0-SURFACE_WAT[cellLoc]/MAC_HEIGHT[cellLoc]))-1.0)) + HP_MAC_MAXROUGH[HAB[cellLoc]],HP_MAC_MINROUGH[HAB[cellLoc]]); 02100 } /* spatial loop end */ 02101 } /* spatial loop end */ 02102 usrErr("Done."); 02103 02104 } /* end of init_eqns */
| void init_canals | ( | int | irun | ) |
Call to initialize the water managment canal network topology and data.
- Parameters:
-
irun a counter of number of times a new simulation run is executed
Definition at line 2110 of file UnitMod.c.
References Canal_Network_Init(), GP_DATUM_DISTANCE, reinitCanals(), SED_ELEV, and usrErr().
02111 { 02112 if (irun == 1) { 02113 usrErr("Initializing Water Management..."); 02114 Canal_Network_Init(GP_DATUM_DISTANCE,SED_ELEV); /* WatMgmt.c - initialize the canal network topology and data */ 02115 usrErr("Done Water Management."); 02116 } 02117 else { 02118 reinitCanals(); 02119 } 02120 02121 }
| void init_succession | ( | void | ) |
Call to initialize the habitat succession module.
Definition at line 2124 of file UnitMod.c.
References HabSwitch_Init().
02125 { 02126 HabSwitch_Init( ); 02127 02128 }

| void reinitBIR | ( | void | ) |
Calls to re-initialize Basin/Indicator-Region data.
Definition at line 2131 of file UnitMod.c.
References BIRbudg_reset(), BIRstats_reset(), Cell_reset_avg(), Cell_reset_hydper(), usrErr(), and usrErr0().
02132 { 02133 usrErr0("Re-initializing Basin/Indicator-Region info..."); 02134 BIRstats_reset(); 02135 BIRbudg_reset(); 02136 Cell_reset_avg(); 02137 Cell_reset_hydper(); 02138 usrErr("Done."); 02139 }
| void reinitCanals | ( | void | ) |
Call to re-initialize canal storages.
Definition at line 2142 of file UnitMod.c.
References CanalReInit(), usrErr(), and usrErr0().
02143 { 02144 usrErr0("Re-initializing Canal depths & constituent masses..."); 02145 CanalReInit(); 02146 usrErr("Done."); 02147 }
| void gen_output | ( | int | step, | |
| ViewParm * | view | |||
| ) |
Generate output.
Create output of spatial maps, point locations and debug data (if any)
- Parameters:
-
step The current iteration number view The struct containing output configuration data
- Remarks:
- You need to ensure that the Outlist_size (define in driver_utilities.h) is greater than (at least equal to) the number of output variables here (important if adding variables to this output list).
- Remarks:
- This UnitMod.c code "fragment" (gen_output function, populating tgen[] array) is generated from the "ModelOutlist_creator_v?.xls" OpenOffice workbook. Editing this source directly is not recommended w/o changing ModelOutlist_creator.
The order of the variables in the Model.outList configuration (model input) file MUST EXACTLY match their order in the tgen[] array of the struct.
The ModelOutlist_creator automatically generates both the Model.outList and the tgen[] code.
It is recommended that you utilize the ModelOutlist_creator to generate both the config file and the source code. NOTE: the code generator has a bug, and puts an 'f' argument in all variables, float or not. You must change doubles ('d') and unsigned chars ('c') by hand.
Definition at line 2162 of file UnitMod.c.
References AIR_TEMP, ALG_INCID_LIGHT, ALG_LIGHT_CF, ALG_LIGHT_EXTINCT, ALG_REFUGE, ALG_SAT, ALG_TEMP_CF, ALG_TOT, ALG_WAT_CF, avgPrint, BCmodel_sfwat, BCmodel_sfwatAvg, C_ALG, C_ALG_AVAIL_MORT, C_ALG_GPP, C_ALG_MORT, C_ALG_MORT_POT, C_ALG_NPP, C_ALG_NUT_CF, C_ALG_PROD_CF, C_ALG_RESP, C_ALG_RESP_POT, C_Peri_mortAvg, C_Peri_nppAvg, C_Peri_PCAvg, C_PeriAvg, C_PeriNutCFAvg, C_PeriRespAvg, CalMonOut, Cell_reset_avg(), Cell_reset_hydper(), outVar_struct::ctype, DAYJUL, DEPOS_ORG_MAT, DOM_DECOMP, DOM_DECOMP_POT, DOM_FR_FLOC, DOM_P_OM, DOM_QUALITY_CF, DOM_SED_AEROB_Z, DOM_SED_ANAEROB_Z, DOM_TEMP_CF, DOM_Z, DOP, DOP_DECOMP, ELEVATION, ETAvg, EvapAvg, FLOC, FLOC_DECOMP, FLOC_DECOMP_POT, FLOC_DECOMP_QUAL_CF, FLOC_DEPO, FLOC_DEPO_POT, FLOC_FR_ALGAE, Floc_fr_phBioAvg, FLOC_Z, FlocP_OMrep, FMOD(), H2O_TEMP, HAB, HYD_DOM_ACTWAT_PRES, HYD_DOM_ACTWAT_VOL, HYD_ET, HYD_EVAP_CALC, HYD_MANNINGS_N, HYD_RCCONDUCT, HYD_SAT_POT_TRANS, HYD_SED_WAT_VOL, HYD_TOT_POT_TRANSP, HYD_TRANSP, HYD_UNSAT_POT_TRANS, HYD_WATER_AVAIL, HydPerAnn, HydRelDepPosNeg, HydRelDepPosNegAvg, HydTotHd, LAI_eff, LAI_effAvg, MAC_HEIGHT, MAC_LAI, MAC_LIGHT_CF, MAC_MAX_BIO, MAC_NOPH_BIOMAS, mac_nph_PC_rep, mac_nph_PCAvg, Mac_nphBioAvg, Mac_nphMortAvg, Mac_nppAvg, MAC_NUT_CF, MAC_PH_BIOMAS, mac_ph_PC_rep, mac_ph_PCAvg, Mac_phBioAvg, Mac_phMortAvg, MAC_PROD_CF, MAC_REL_BIOM, MAC_SALT_CF, MAC_TEMP_CF, MAC_TOT_BIOM, Mac_totBioAvg, MAC_WATER_AVAIL_CF, MAC_WATER_CF, MacNutCfAvg, MacWatCfAvg, Manning_nAvg, NC_ALG, NC_ALG_AVAIL_MORT, NC_ALG_GPP, NC_ALG_MORT, NC_ALG_MORT_POT, NC_ALG_NPP, NC_ALG_NUT_CF, NC_ALG_PROD_CF, NC_ALG_RESP, NC_ALG_RESP_POT, NC_Peri_mortAvg, NC_Peri_nppAvg, NC_Peri_PCAvg, NC_PeriAvg, NC_PeriNutCFAvg, NC_PeriRespAvg, NPHBIO_AVAIL, NPHBIO_MORT, NPHBIO_MORT_POT, NPHBIO_REFUGE, NPHBIO_SAT, NPHBIO_TRANSLOC, NPHBIO_TRANSLOC_POT, numOutputs, ON_MAP, P_SUM_CELL, PeriAvg, PeriLiteCFAvg, outVar_struct::pfvar, PHBIO_AVAIL, PHBIO_MORT, PHBIO_MORT_POT, PHBIO_NPP, PHBIO_REFUGE, PHBIO_SAT, PHBIO_TRANSLOC, PHBIO_TRANSLOC_POT, outVar_struct::pname, RainAvg, SAL_SED_WT, SAL_SF_WT, SALT_Atm_Depos, SALT_SED_TO_SF_FLOW, SALT_SED_WT, SALT_SFWAT_DOWNFL, SALT_SFWAT_DOWNFL_POT, SALT_SURF_WT, SaltSedAvg, SaltSfAvg, SAT_TO_UNSAT_FL, SAT_VS_UNSAT, SAT_WATER, SAT_WT_HEAD, SAT_WT_RECHG, SAT_WT_TRANSP, SED_ELEV, SED_INACT_Z, SedElevAvg, SF_WT_EVAP, SF_WT_FROM_RAIN, SF_WT_INFILTRATION, SF_WT_POT_INF, SF_WT_TO_SAT_DOWNFLOW, SF_WT_VEL_mag, SF_WT_VEL_magAvg, SfWatAvg, SFWT_VOL, soil_MOIST_CF, SOLRAD274, SOLRADGRD, viewParm::step, SURFACE_WAT, TotHeadAvg, TP_Act_to_TotRep, TP_Atm_Depos, TP_DNFLOW, TP_DNFLOW_POT, TP_K, TP_SED_CONC, TP_SED_MINER, TP_SED_WT, TP_SED_WT_AZ, TP_SEDWT_CONCACT, TP_SEDWT_CONCACTMG, TP_SEDWT_UPTAKE, TP_settl, TP_settlAvg, TP_settlDays, TP_SF_WT, TP_SFWT_CONC, TP_SFWT_CONC_MG, TP_SFWT_MINER, TP_SFWT_UPTAK, TP_SORB, TP_SORB_POT, TP_SORBCONC, TP_SORBCONC_rep, TP_SORBTION, TP_UPFLOW, TP_UPFLOW_POT, TPSedMinAvg, TPSedUptAvg, TPSedWatAvg, TPSfMinAvg, TPSfUptAvg, TPSfWatAvg, TPSorbAvg, TPtoSOIL_rep, TPtoSOILAvg, TPtoVOLAvg, TranspAvg, UNSAT_AVAIL, UNSAT_CAP, UNSAT_DEPTH, UNSAT_HYD_COND_CF, UNSAT_MOIST_PRP, UNSAT_PERC, UNSAT_TO_SAT_FL, UNSAT_TRANSP, UNSAT_WATER, UNSAT_WT_POT, UnsatMoistAvg, UnsatZavg, and write_output().
02163 { 02164 #define numOutputs 50000 02165 static int iw[numOutputs]; 02166 int oIndex; 02167 ViewParm *vp; 02168 02176 static outVar_struct tgen[] = { 02177 { (float**)&TP_settlDays, "TP_settlDays", 'f' }, 02178 { (float**)&FLOC, "FLOC", 'f' }, 02179 { (float**)&FLOC_DECOMP, "FLOC_DECOMP", 'f' }, 02180 { (float**)&FLOC_DECOMP_POT, "FLOC_DECOMP_POT", 'f' }, 02181 { (float**)&FLOC_DECOMP_QUAL_CF, "FLOC_DECOMP_QUAL_CF", 'f' }, 02182 { (float**)&FLOC_DEPO, "FLOC_DEPO", 'f' }, 02183 { (float**)&FLOC_DEPO_POT, "FLOC_DEPO_POT", 'f' }, 02184 { (float**)&FLOC_FR_ALGAE, "FLOC_FR_ALGAE", 'f' }, 02185 { (float**)&FLOC_Z, "FLOC_Z", 'f' }, 02186 { (float**)&FlocP_OMrep, "FlocP_OMrep", 'f' }, 02187 { (float**)&soil_MOIST_CF, "soil_MOIST_CF", 'f' }, 02188 { (float**)&TP_SED_MINER, "TP_SED_MINER", 'f' }, 02189 { (float**)&TP_SFWT_MINER, "TP_SFWT_MINER", 'f' }, 02190 { (float**)&AIR_TEMP, "AIR_TEMP", 'f' }, 02191 { (unsigned char**)&HAB, "HAB", 'c' }, 02192 { (float**)&SOLRAD274, "SOLRAD274", 'f' }, 02193 { (float**)&SOLRADGRD, "SOLRADGRD", 'f' }, 02194 { (float**)&H2O_TEMP, "H2O_TEMP", 'f' }, 02195 { (float**)&HYD_DOM_ACTWAT_PRES, "HYD_DOM_ACTWAT_PRES", 'f' }, 02196 { (float**)&HYD_DOM_ACTWAT_VOL, "HYD_DOM_ACTWAT_VOL", 'f' }, 02197 { (float**)&HYD_ET, "HYD_ET", 'f' }, 02198 { (float**)&HYD_EVAP_CALC, "HYD_EVAP_CALC", 'f' }, 02199 { (float**)&HYD_MANNINGS_N, "HYD_MANNINGS_N", 'f' }, 02200 { (float**)&HYD_SAT_POT_TRANS, "HYD_SAT_POT_TRANS", 'f' }, 02201 { (float**)&HYD_SED_WAT_VOL, "HYD_SED_WAT_VOL", 'f' }, 02202 { (float**)&HYD_TOT_POT_TRANSP, "HYD_TOT_POT_TRANSP", 'f' }, 02203 { (float**)&HYD_TRANSP, "HYD_TRANSP", 'f' }, 02204 { (float**)&HYD_UNSAT_POT_TRANS, "HYD_UNSAT_POT_TRANS", 'f' }, 02205 { (float**)&HYD_WATER_AVAIL, "HYD_WATER_AVAIL", 'f' }, 02206 { (float**)&HydTotHd, "HydTotHd", 'f' }, 02207 { (float**)&HydRelDepPosNeg, "HydRelDepPosNeg", 'f' }, 02208 { (float**)&LAI_eff, "LAI_eff", 'f' }, 02209 { (float**)&MAC_WATER_AVAIL_CF, "MAC_WATER_AVAIL_CF", 'f' }, 02210 { (float**)&SAT_TO_UNSAT_FL, "SAT_TO_UNSAT_FL", 'f' }, 02211 { (float**)&SAT_VS_UNSAT, "SAT_VS_UNSAT", 'f' }, 02212 { (float**)&SAT_WATER, "SAT_WATER", 'f' }, 02213 { (float**)&SAT_WT_HEAD, "SAT_WT_HEAD", 'f' }, 02214 { (float**)&SAT_WT_RECHG, "SAT_WT_RECHG", 'f' }, 02215 { (float**)&SAT_WT_TRANSP, "SAT_WT_TRANSP", 'f' }, 02216 { (float**)&SF_WT_EVAP, "SF_WT_EVAP", 'f' }, 02217 { (float**)&SF_WT_FROM_RAIN, "SF_WT_FROM_RAIN", 'f' }, 02218 { (float**)&SF_WT_INFILTRATION, "SF_WT_INFILTRATION", 'f' }, 02219 { (float**)&SF_WT_POT_INF, "SF_WT_POT_INF", 'f' }, 02220 { (float**)&SF_WT_TO_SAT_DOWNFLOW, "SF_WT_TO_SAT_DOWNFLOW", 'f' }, 02221 { (float**)&SF_WT_VEL_mag, "SF_WT_VEL_mag", 'f' }, 02222 { (float**)&BCmodel_sfwat, "BCmodel_sfwat", 'f' }, 02223 { (float**)&SFWT_VOL, "SFWT_VOL", 'f' }, 02224 { (float**)&SURFACE_WAT, "SURFACE_WAT", 'f' }, 02225 { (float**)&UNSAT_AVAIL, "UNSAT_AVAIL", 'f' }, 02226 { (float**)&UNSAT_CAP, "UNSAT_CAP", 'f' }, 02227 { (float**)&UNSAT_DEPTH, "UNSAT_DEPTH", 'f' }, 02228 { (float**)&UNSAT_HYD_COND_CF, "UNSAT_HYD_COND_CF", 'f' }, 02229 { (float**)&UNSAT_MOIST_PRP, "UNSAT_MOIST_PRP", 'f' }, 02230 { (float**)&UNSAT_PERC, "UNSAT_PERC", 'f' }, 02231 { (float**)&UNSAT_TO_SAT_FL, "UNSAT_TO_SAT_FL", 'f' }, 02232 { (float**)&UNSAT_TRANSP, "UNSAT_TRANSP", 'f' }, 02233 { (float**)&UNSAT_WATER, "UNSAT_WATER", 'f' }, 02234 { (float**)&UNSAT_WT_POT, "UNSAT_WT_POT", 'f' }, 02235 { (float**)&ELEVATION, "ELEVATION", 'f' }, 02236 { (float**)&HYD_RCCONDUCT, "HYD_RCCONDUCT", 'f' }, 02237 { (unsigned char**)&ON_MAP, "ON_MAP", 'c' }, 02238 { (float**)&SED_INACT_Z, "SED_INACT_Z", 'f' }, 02239 { (float**)&MAC_HEIGHT, "MAC_HEIGHT", 'f' }, 02240 { (float**)&MAC_LAI, "MAC_LAI", 'f' }, 02241 { (float**)&MAC_LIGHT_CF, "MAC_LIGHT_CF", 'f' }, 02242 { (float**)&MAC_MAX_BIO, "MAC_MAX_BIO", 'f' }, 02243 { (float**)&MAC_NOPH_BIOMAS, "MAC_NOPH_BIOMAS", 'f' }, 02244 { (float**)&mac_nph_PC_rep, "mac_nph_PC_rep", 'f' }, 02245 { (float**)&MAC_NUT_CF, "MAC_NUT_CF", 'f' }, 02246 { (float**)&MAC_PH_BIOMAS, "MAC_PH_BIOMAS", 'f' }, 02247 { (float**)&mac_ph_PC_rep, "mac_ph_PC_rep", 'f' }, 02248 { (float**)&MAC_PROD_CF, "MAC_PROD_CF", 'f' }, 02249 { (float**)&MAC_REL_BIOM, "MAC_REL_BIOM", 'f' }, 02250 { (float**)&MAC_SALT_CF, "MAC_SALT_CF", 'f' }, 02251 { (float**)&MAC_TEMP_CF, "MAC_TEMP_CF", 'f' }, 02252 { (float**)&MAC_TOT_BIOM, "MAC_TOT_BIOM", 'f' }, 02253 { (float**)&MAC_WATER_CF, "MAC_WATER_CF", 'f' }, 02254 { (float**)&NPHBIO_AVAIL, "NPHBIO_AVAIL", 'f' }, 02255 { (float**)&NPHBIO_MORT, "NPHBIO_MORT", 'f' }, 02256 { (float**)&NPHBIO_MORT_POT, "NPHBIO_MORT_POT", 'f' }, 02257 { (float**)&NPHBIO_REFUGE, "NPHBIO_REFUGE", 'f' }, 02258 { (float**)&NPHBIO_SAT, "NPHBIO_SAT", 'f' }, 02259 { (float**)&NPHBIO_TRANSLOC, "NPHBIO_TRANSLOC", 'f' }, 02260 { (float**)&NPHBIO_TRANSLOC_POT, "NPHBIO_TRANSLOC_POT", 'f' }, 02261 { (float**)&PHBIO_AVAIL, "PHBIO_AVAIL", 'f' }, 02262 { (float**)&PHBIO_MORT, "PHBIO_MORT", 'f' }, 02263 { (float**)&PHBIO_MORT_POT, "PHBIO_MORT_POT", 'f' }, 02264 { (float**)&PHBIO_NPP, "PHBIO_NPP", 'f' }, 02265 { (float**)&PHBIO_REFUGE, "PHBIO_REFUGE", 'f' }, 02266 { (float**)&PHBIO_SAT, "PHBIO_SAT", 'f' }, 02267 { (float**)&PHBIO_TRANSLOC, "PHBIO_TRANSLOC", 'f' }, 02268 { (float**)&PHBIO_TRANSLOC_POT, "PHBIO_TRANSLOC_POT", 'f' }, 02269 { (float**)&TP_SEDWT_UPTAKE, "TP_SEDWT_UPTAKE", 'f' }, 02270 { (float**)&ALG_INCID_LIGHT, "ALG_INCID_LIGHT", 'f' }, 02271 { (float**)&ALG_LIGHT_CF, "ALG_LIGHT_CF", 'f' }, 02272 { (float**)&ALG_LIGHT_EXTINCT, "ALG_LIGHT_EXTINCT", 'f' }, 02273 { (float**)&ALG_REFUGE, "ALG_REFUGE", 'f' }, 02274 { (float**)&ALG_SAT, "ALG_SAT", 'f' }, 02275 { (float**)&ALG_TEMP_CF, "ALG_TEMP_CF", 'f' }, 02276 { (float**)&ALG_TOT, "ALG_TOT", 'f' }, 02277 { (float**)&ALG_WAT_CF, "ALG_WAT_CF", 'f' }, 02278 { (float**)&C_ALG, "C_ALG", 'f' }, 02279 { (float**)&C_ALG_AVAIL_MORT, "C_ALG_AVAIL_MORT", 'f' }, 02280 { (float**)&C_ALG_GPP, "C_ALG_GPP", 'f' }, 02281 { (float**)&C_ALG_MORT, "C_ALG_MORT", 'f' }, 02282 { (float**)&C_ALG_MORT_POT, "C_ALG_MORT_POT", 'f' }, 02283 { (float**)&C_ALG_NPP, "C_ALG_NPP", 'f' }, 02284 { (float**)&C_ALG_NUT_CF, "C_ALG_NUT_CF", 'f' }, 02285 { (float**)&C_ALG_PROD_CF, "C_ALG_PROD_CF", 'f' }, 02286 { (float**)&C_ALG_RESP, "C_ALG_RESP", 'f' }, 02287 { (float**)&C_ALG_RESP_POT, "C_ALG_RESP_POT", 'f' }, 02288 { (float**)&NC_ALG, "NC_ALG", 'f' }, 02289 { (float**)&NC_ALG_AVAIL_MORT, "NC_ALG_AVAIL_MORT", 'f' }, 02290 { (float**)&NC_ALG_GPP, "NC_ALG_GPP", 'f' }, 02291 { (float**)&NC_ALG_MORT, "NC_ALG_MORT", 'f' }, 02292 { (float**)&NC_ALG_MORT_POT, "NC_ALG_MORT_POT", 'f' }, 02293 { (float**)&NC_ALG_NPP, "NC_ALG_NPP", 'f' }, 02294 { (float**)&NC_ALG_NUT_CF, "NC_ALG_NUT_CF", 'f' }, 02295 { (float**)&NC_ALG_PROD_CF, "NC_ALG_PROD_CF", 'f' }, 02296 { (float**)&NC_ALG_RESP, "NC_ALG_RESP", 'f' }, 02297 { (float**)&NC_ALG_RESP_POT, "NC_ALG_RESP_POT", 'f' }, 02298 { (float**)&TP_SFWT_UPTAK, "TP_SFWT_UPTAK", 'f' }, 02299 { (float**)&TP_Act_to_TotRep, "TP_Act_to_TotRep", 'f' }, 02300 { (float**)&TP_DNFLOW, "TP_DNFLOW", 'f' }, 02301 { (float**)&TP_DNFLOW_POT, "TP_DNFLOW_POT", 'f' }, 02302 { (float**)&TP_Atm_Depos, "TP_Atm_Depos", 'f' }, 02303 { (float**)&TP_K, "TP_K", 'f' }, 02304 { (double**)&TP_SED_CONC, "TP_SED_CONC", 'd' }, 02305 { (double**)&TP_SED_WT, "TP_SED_WT", 'd' }, 02306 { (double**)&TP_SED_WT_AZ, "TP_SED_WT_AZ", 'd' }, 02307 { (double**)&TP_SEDWT_CONCACT, "TP_SEDWT_CONCACT", 'd' }, 02308 { (float**)&TP_SEDWT_CONCACTMG, "TP_SEDWT_CONCACTMG", 'f' }, 02309 { (float**)&TP_settl, "TP_settl", 'f' }, 02310 { (double**)&TP_SF_WT, "TP_SF_WT", 'd' }, 02311 { (double**)&TP_SFWT_CONC, "TP_SFWT_CONC", 'd' }, 02312 { (float**)&TP_SFWT_CONC_MG, "TP_SFWT_CONC_MG", 'f' }, 02313 { (double**)&TP_SORB, "TP_SORB", 'd' }, 02314 { (float**)&TP_SORB_POT, "TP_SORB_POT", 'f' }, 02315 { (double**)&TP_SORBCONC, "TP_SORBCONC", 'd' }, 02316 { (float**)&TP_SORBCONC_rep, "TP_SORBCONC_rep", 'f' }, 02317 { (float**)&TP_SORBTION, "TP_SORBTION", 'f' }, 02318 { (float**)&TP_UPFLOW, "TP_UPFLOW", 'f' }, 02319 { (float**)&TP_UPFLOW_POT, "TP_UPFLOW_POT", 'f' }, 02320 { (double**)&SAL_SED_WT, "SAL_SED_WT", 'd' }, 02321 { (float**)&SAL_SF_WT, "SAL_SF_WT", 'f' }, 02322 { (float**)&SALT_SED_TO_SF_FLOW, "SALT_SED_TO_SF_FLOW", 'f' }, 02323 { (double**)&SALT_SED_WT, "SALT_SED_WT", 'd' }, 02324 { (float**)&SALT_SFWAT_DOWNFL, "SALT_SFWAT_DOWNFL", 'f' }, 02325 { (float**)&SALT_SFWAT_DOWNFL_POT, "SALT_SFWAT_DOWNFL_POT", 'f' }, 02326 { (double**)&SALT_SURF_WT, "SALT_SURF_WT", 'd' }, 02327 { (float**)&SALT_Atm_Depos, "SALT_Atm_Depos", 'f' }, 02328 { (double**)&DEPOS_ORG_MAT, "DEPOS_ORG_MAT", 'd' }, 02329 { (float**)&DOM_DECOMP, "DOM_DECOMP", 'f' }, 02330 { (float**)&DOM_DECOMP_POT, "DOM_DECOMP_POT", 'f' }, 02331 { (float**)&DOM_FR_FLOC, "DOM_FR_FLOC", 'f' }, 02332 { (double**)&DOM_P_OM, "DOM_P_OM", 'd' }, 02333 { (float**)&DOM_QUALITY_CF, "DOM_QUALITY_CF", 'f' }, 02334 { (float**)&DOM_SED_AEROB_Z, "DOM_SED_AEROB_Z", 'f' }, 02335 { (float**)&DOM_SED_ANAEROB_Z, "DOM_SED_ANAEROB_Z", 'f' }, 02336 { (float**)&DOM_TEMP_CF, "DOM_TEMP_CF", 'f' }, 02337 { (float**)&DOM_Z, "DOM_Z", 'f' }, 02338 { (double**)&DOP, "DOP", 'd' }, 02339 { (float**)&DOP_DECOMP, "DOP_DECOMP", 'f' }, 02340 { (float**)&P_SUM_CELL, "P_SUM_CELL", 'f' }, 02341 { (float**)&SED_ELEV, "SED_ELEV", 'f' }, 02342 { (float**)&TPtoSOIL_rep, "TPtoSOIL_rep", 'f' }, 02343 { (float**)&Floc_fr_phBioAvg, "Floc_fr_phBioAvg", 'f' }, 02344 { (float**)&TPSfMinAvg, "TPSfMinAvg", 'f' }, 02345 { (float**)&ETAvg, "ETAvg", 'f' }, 02346 { (float**)&EvapAvg, "EvapAvg", 'f' }, 02347 { (float**)&HydPerAnn, "HydPerAnn", 'f' }, 02348 { (float**)&LAI_effAvg, "LAI_effAvg", 'f' }, 02349 { (float**)&Manning_nAvg, "Manning_nAvg", 'f' }, 02350 { (float**)&RainAvg, "RainAvg", 'f' }, 02351 { (float**)&SfWatAvg, "SfWatAvg", 'f' }, 02352 { (float**)&TotHeadAvg, "TotHeadAvg", 'f' }, 02353 { (float**)&HydRelDepPosNegAvg, "HydRelDepPosNegAvg", 'f' }, 02354 { (float**)&TranspAvg, "TranspAvg", 'f' }, 02355 { (float**)&UnsatMoistAvg, "UnsatMoistAvg", 'f' }, 02356 { (float**)&UnsatZavg, "UnsatZavg", 'f' }, 02357 { (float**)&SF_WT_VEL_magAvg, "SF_WT_VEL_magAvg", 'f' }, 02358 { (float**)&BCmodel_sfwatAvg, "BCmodel_sfwatAvg", 'f' }, 02359 { (float**)&mac_nph_PCAvg, "mac_nph_PCAvg", 'f' }, 02360 { (float**)&Mac_nphBioAvg, "Mac_nphBioAvg", 'f' }, 02361 { (float**)&Mac_nphMortAvg, "Mac_nphMortAvg", 'f' }, 02362 { (float**)&Mac_nppAvg, "Mac_nppAvg", 'f' }, 02363 { (float**)&mac_ph_PCAvg, "mac_ph_PCAvg", 'f' }, 02364 { (float**)&Mac_phBioAvg, "Mac_phBioAvg", 'f' }, 02365 { (float**)&Mac_phMortAvg, "Mac_phMortAvg", 'f' }, 02366 { (float**)&Mac_totBioAvg, "Mac_totBioAvg", 'f' }, 02367 { (float**)&MacNutCfAvg, "MacNutCfAvg", 'f' }, 02368 { (float**)&MacWatCfAvg, "MacWatCfAvg", 'f' }, 02369 { (float**)&TPSedUptAvg, "TPSedUptAvg", 'f' }, 02370 { (float**)&C_Peri_mortAvg, "C_Peri_mortAvg", 'f' }, 02371 { (float**)&C_Peri_nppAvg, "C_Peri_nppAvg", 'f' }, 02372 { (float**)&C_Peri_PCAvg, "C_Peri_PCAvg", 'f' }, 02373 { (float**)&C_PeriAvg, "C_PeriAvg", 'f' }, 02374 { (float**)&C_PeriNutCFAvg, "C_PeriNutCFAvg", 'f' }, 02375 { (float**)&C_PeriRespAvg, "C_PeriRespAvg", 'f' }, 02376 { (float**)&NC_Peri_mortAvg, "NC_Peri_mortAvg", 'f' }, 02377 { (float**)&NC_Peri_nppAvg, "NC_Peri_nppAvg", 'f' }, 02378 { (float**)&NC_Peri_PCAvg, "NC_Peri_PCAvg", 'f' }, 02379 { (float**)&NC_PeriAvg, "NC_PeriAvg", 'f' }, 02380 { (float**)&NC_PeriNutCFAvg, "NC_PeriNutCFAvg", 'f' }, 02381 { (float**)&NC_PeriRespAvg, "NC_PeriRespAvg", 'f' }, 02382 { (float**)&PeriAvg, "PeriAvg", 'f' }, 02383 { (float**)&PeriLiteCFAvg, "PeriLiteCFAvg", 'f' }, 02384 { (float**)&TPSfUptAvg, "TPSfUptAvg", 'f' }, 02385 { (float**)&TP_settlAvg, "TP_settlAvg", 'f' }, 02386 { (float**)&TPSedWatAvg, "TPSedWatAvg", 'f' }, 02387 { (float**)&TPSfWatAvg, "TPSfWatAvg", 'f' }, 02388 { (float**)&SaltSedAvg, "SaltSedAvg", 'f' }, 02389 { (float**)&SaltSfAvg, "SaltSfAvg", 'f' }, 02390 { (float**)&SedElevAvg, "SedElevAvg", 'f' }, 02391 { (float**)&TPSedMinAvg, "TPSedMinAvg", 'f' }, 02392 { (float**)&TPSorbAvg, "TPSorbAvg", 'f' }, 02393 { (float**)&TPtoSOILAvg, "TPtoSOILAvg", 'f' }, 02394 { (float**)&TPtoVOLAvg, "TPtoVOLAvg", 'f' }, 02395 02396 02397 02398 02399 02400 02401 02402 02403 { NULL, NULL, '\0' }, 02404 }; 02405 02406 outVar_struct *ptable; 02407 int i; 02408 02409 for (i = 0, ptable = tgen; ptable->pfvar != NULL; ptable++, i++) 02410 { 02411 vp = view + i; 02412 02413 /* TODO: develop flexible output of calendar-based and julian-based outsteps (jan 11, 2005) */ 02414 if (vp->step > 0) 02415 02416 if (strcmp(ptable->pname,"HydPerAnn")!=0) { /* i.e., not the HydPerAnn variable */ 02417 02418 if (step % vp->step == 0 && (vp->step != CalMonOut) ) { /* standard julian-day outstep interval variables (note: the != CalMonOut needed for step=0) */ 02419 oIndex = iw[i]++; 02420 write_output(oIndex, vp, *(ptable->pfvar), ptable->pname, ptable->ctype, step); 02421 } 02422 else if ( (avgPrint) && (vp->step == CalMonOut) ) { /* variables output at the special 1-calendar-month outstep interval */ 02423 oIndex = iw[i]++; 02424 write_output(oIndex, vp, *(ptable->pfvar), ptable->pname, ptable->ctype, step); 02425 } 02426 } 02427 02428 else 02429 if (FMOD(DAYJUL, 273.0) ==0) { /* hydroperiod is printed at a special-case time (approximately Oct 1 every year) */ 02430 oIndex = iw[i]++; 02431 write_output(oIndex, vp, *(ptable->pfvar), ptable->pname, ptable->ctype, step); 02432 } 02433 02434 02435 } 02436 02437 /* after printing, zero the arrays holding averages or hydroperiods (v2.2.1note using avgPrint var)*/ 02438 if (avgPrint) { 02439 Cell_reset_avg(); 02440 } 02441 02442 if (FMOD(DAYJUL, 273.0) ==0) { 02443 Cell_reset_hydper(); 02444 } 02445 02446 02447 } /* end of gen_output routine */
| void ReadGlobalParms | ( | char * | s_parm_name, | |
| int | s_parm_relval | |||
| ) |
Acquire the model parameters that are global to the domain.
- Parameters:
-
s_parm_name Name of the sensitivity parameter being varied for this run s_parm_relval Indicator of current value of relative range in sensitivity parm values: nominal (0), low (1), or high (2)
Definition at line 2532 of file UnitMod.c.
References get_global_parm(), GP_alg_alkP_min, GP_ALG_C_TO_OM, GP_ALG_IC_MULT, GP_alg_light_ext_coef, GP_ALG_LIGHT_SAT, GP_ALG_PC, GP_alg_R_accel, GP_ALG_RC_MORT, GP_ALG_RC_MORT_DRY, GP_ALG_RC_PROD, GP_ALG_RC_RESP, GP_ALG_REF_MULT, GP_ALG_SHADE_FACTOR, GP_ALG_TEMP_OPT, GP_alg_uptake_coef, GP_AlgComp, GP_algMortDepth, GP_ALTIT, GP_C_ALG_KS_P, GP_C_ALG_threshTP, GP_calibDecomp, GP_calibET, GP_calibGWat, GP_CL_IN_RAIN, GP_DATUM_DISTANCE, GP_DetentZ, GP_dispLenRef, GP_dispParm, GP_DOM_decomp_coef, GP_DOM_DECOMP_POPT, GP_DOM_DECOMP_TOPT, GP_DOM_DECOMPRED, GP_DOM_RCDECOMP, GP_Floc_BD, GP_Floc_rcSoil, GP_FlocMax, GP_HYD_IC_SFWAT_ADD, GP_HYD_IC_UNSAT_ADD, GP_HYD_ICUNSATMOIST, GP_HYD_RCRECHG, GP_IC_BATHY_MULT, GP_IC_BulkD_MULT, GP_IC_DOM_BD_MULT, GP_IC_ELEV_MULT, GP_IC_TPtoSOIL_MULT, GP_IDW_pow, GP_LATDEG, GP_MAC_IC_MULT, GP_MAC_REFUG_MULT, GP_mac_uptake_coef, GP_mann_height_coef, GP_mannDepthPow, GP_mannHeadPow, GP_MinCheck, GP_NC_ALG_KS_P, GP_PO4toTP, GP_PO4toTPint, GP_settlVel, GP_SLRise, GP_SOLOMEGA, GP_sorbToTP, GP_TP_DIFFCOEF, GP_TP_DIFFDEPTH, GP_TP_ICSEDWAT, GP_TP_ICSFWAT, GP_TP_IN_RAIN, GP_TP_K_INTER, GP_TP_K_SLOPE, GP_TP_P_OM, GP_TPpart_thresh, GP_WQMthresh, and GP_WQualMonitZ.
02533 { 02534 02535 /* Geography, hydrology */ 02536 GP_SOLOMEGA = get_global_parm(s_parm_name, s_parm_relval,"GP_SOLOMEGA"); 02537 GP_ALTIT = get_global_parm(s_parm_name, s_parm_relval,"GP_ALTIT"); 02538 GP_LATDEG = get_global_parm(s_parm_name, s_parm_relval,"GP_LATDEG"); 02539 GP_mannDepthPow = get_global_parm(s_parm_name, s_parm_relval,"GP_mannDepthPow"); 02540 GP_mannHeadPow = get_global_parm(s_parm_name, s_parm_relval,"GP_mannHeadPow"); 02541 GP_calibGWat = get_global_parm(s_parm_name, s_parm_relval,"GP_calibGWat"); 02542 GP_IDW_pow = get_global_parm(s_parm_name, s_parm_relval,"GP_IDW_pow"); 02543 GP_calibET = get_global_parm(s_parm_name, s_parm_relval,"GP_calibET"); 02544 GP_DATUM_DISTANCE = get_global_parm(s_parm_name, s_parm_relval,"GP_DATUM_DISTANCE"); 02545 GP_HYD_IC_SFWAT_ADD = get_global_parm(s_parm_name, s_parm_relval,"GP_HYD_IC_SFWAT_ADD"); 02546 GP_HYD_IC_UNSAT_ADD = get_global_parm(s_parm_name, s_parm_relval,"GP_HYD_IC_UNSAT_ADD"); 02547 GP_HYD_RCRECHG = get_global_parm(s_parm_name, s_parm_relval,"GP_HYD_RCRECHG"); 02548 GP_HYD_ICUNSATMOIST = get_global_parm(s_parm_name, s_parm_relval,"GP_HYD_ICUNSATMOIST"); 02549 GP_DetentZ = get_global_parm(s_parm_name, s_parm_relval,"GP_DetentZ"); 02550 GP_WQualMonitZ = get_global_parm(s_parm_name, s_parm_relval,"GP_WQualMonitZ"); 02551 GP_MinCheck = get_global_parm(s_parm_name, s_parm_relval,"GP_MinCheck"); 02552 GP_SLRise = get_global_parm(s_parm_name, s_parm_relval,"GP_SLRise"); 02553 GP_dispLenRef = get_global_parm(s_parm_name, s_parm_relval,"GP_dispLenRef"); 02554 GP_dispParm = get_global_parm(s_parm_name, s_parm_relval,"GP_dispParm"); 02555 02556 /* Periphyton/Algae */ 02557 GP_ALG_IC_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_IC_MULT"); 02558 GP_alg_uptake_coef = get_global_parm(s_parm_name, s_parm_relval,"GP_alg_uptake_coef"); 02559 GP_ALG_SHADE_FACTOR = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_SHADE_FACTOR"); 02560 GP_algMortDepth = get_global_parm(s_parm_name, s_parm_relval,"GP_algMortDepth"); 02561 GP_ALG_RC_MORT_DRY = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_RC_MORT_DRY"); 02562 GP_ALG_RC_MORT = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_RC_MORT"); 02563 GP_ALG_RC_PROD = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_RC_PROD"); 02564 GP_ALG_RC_RESP = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_RC_RESP"); 02565 GP_alg_R_accel = get_global_parm(s_parm_name, s_parm_relval,"GP_alg_R_accel"); 02566 GP_AlgComp = get_global_parm(s_parm_name, s_parm_relval,"GP_AlgComp"); 02567 GP_ALG_REF_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_REF_MULT"); 02568 GP_NC_ALG_KS_P = get_global_parm(s_parm_name, s_parm_relval,"GP_NC_ALG_KS_P"); 02569 GP_alg_alkP_min = get_global_parm(s_parm_name, s_parm_relval,"GP_alg_alkP_min"); 02570 GP_C_ALG_KS_P = get_global_parm(s_parm_name, s_parm_relval,"GP_C_ALG_KS_P"); 02571 GP_ALG_TEMP_OPT = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_TEMP_OPT"); 02572 GP_C_ALG_threshTP = get_global_parm(s_parm_name, s_parm_relval,"GP_C_ALG_threshTP"); 02573 GP_ALG_C_TO_OM = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_C_TO_OM"); 02574 GP_alg_light_ext_coef = get_global_parm(s_parm_name, s_parm_relval,"GP_alg_light_ext_coef"); 02575 GP_ALG_LIGHT_SAT = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_LIGHT_SAT"); 02576 GP_ALG_PC = get_global_parm(s_parm_name, s_parm_relval,"GP_ALG_PC"); 02577 02578 /* Soil */ 02579 GP_DOM_RCDECOMP = get_global_parm(s_parm_name, s_parm_relval,"GP_DOM_RCDECOMP"); 02580 GP_DOM_DECOMPRED = get_global_parm(s_parm_name, s_parm_relval,"GP_DOM_DECOMPRED"); 02581 GP_calibDecomp = get_global_parm(s_parm_name, s_parm_relval,"GP_calibDecomp"); 02582 GP_DOM_decomp_coef = get_global_parm(s_parm_name, s_parm_relval,"GP_DOM_decomp_coef"); 02583 GP_DOM_DECOMP_POPT = get_global_parm(s_parm_name, s_parm_relval,"GP_DOM_DECOMP_POPT"); 02584 GP_DOM_DECOMP_TOPT = get_global_parm(s_parm_name, s_parm_relval,"GP_DOM_DECOMP_TOPT"); 02585 GP_sorbToTP = get_global_parm(s_parm_name, s_parm_relval,"GP_sorbToTP"); 02586 GP_IC_ELEV_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_IC_ELEV_MULT"); 02587 GP_IC_BATHY_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_IC_BATHY_MULT"); 02588 GP_IC_DOM_BD_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_IC_DOM_BD_MULT"); 02589 GP_IC_BulkD_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_IC_BulkD_MULT"); 02590 GP_IC_TPtoSOIL_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_IC_TPtoSOIL_MULT"); 02591 02592 /* Macrophytes */ 02593 GP_MAC_IC_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_MAC_IC_MULT"); 02594 GP_MAC_REFUG_MULT = get_global_parm(s_parm_name, s_parm_relval,"GP_MAC_REFUG_MULT"); 02595 GP_mac_uptake_coef = get_global_parm(s_parm_name, s_parm_relval,"GP_mac_uptake_coef"); 02596 GP_mann_height_coef = get_global_parm(s_parm_name, s_parm_relval,"GP_mann_height_coef"); 02597 02598 /* Floc */ 02599 GP_Floc_BD = get_global_parm(s_parm_name, s_parm_relval,"GP_Floc_BD"); 02600 GP_FlocMax = get_global_parm(s_parm_name, s_parm_relval,"GP_FlocMax"); 02601 GP_TP_P_OM = get_global_parm(s_parm_name, s_parm_relval,"GP_TP_P_OM"); 02602 GP_Floc_rcSoil = get_global_parm(s_parm_name, s_parm_relval,"GP_Floc_rcSoil"); 02603 02604 /* Phosphorus */ 02605 GP_TP_DIFFCOEF = get_global_parm(s_parm_name, s_parm_relval,"GP_TP_DIFFCOEF"); 02606 GP_TP_K_INTER = get_global_parm(s_parm_name, s_parm_relval,"GP_TP_K_INTER"); 02607 GP_TP_K_SLOPE = get_global_parm(s_parm_name, s_parm_relval,"GP_TP_K_SLOPE"); 02608 GP_WQMthresh = get_global_parm(s_parm_name, s_parm_relval,"GP_WQMthresh"); 02609 GP_PO4toTP = get_global_parm(s_parm_name, s_parm_relval,"GP_PO4toTP"); 02610 GP_TP_IN_RAIN = get_global_parm(s_parm_name, s_parm_relval,"GP_TP_IN_RAIN"); 02611 GP_CL_IN_RAIN = get_global_parm(s_parm_name, s_parm_relval,"GP_CL_IN_RAIN"); 02612 GP_PO4toTPint = get_global_parm(s_parm_name, s_parm_relval,"GP_PO4toTPint"); 02613 GP_TP_ICSFWAT = get_global_parm(s_parm_name, s_parm_relval,"GP_TP_ICSFWAT"); 02614 GP_TP_ICSEDWAT = get_global_parm(s_parm_name, s_parm_relval,"GP_TP_ICSEDWAT"); 02615 GP_TPpart_thresh = get_global_parm(s_parm_name, s_parm_relval,"GP_TPpart_thresh"); 02616 GP_TP_DIFFDEPTH = get_global_parm(s_parm_name, s_parm_relval,"GP_TP_DIFFDEPTH"); 02617 GP_settlVel = get_global_parm(s_parm_name, s_parm_relval,"GP_settlVel"); 02618 02619 }

| void ReadHabParms | ( | char * | s_parm_name, | |
| int | s_parm_relval | |||
| ) |
Acquire the model parameters that are specific to different habitat types in the domain.
- Parameters:
-
s_parm_name Name of the sensitivity parameter being varied for this run s_parm_relval Indicator of current value of relative range in sensitivity parm values: nominal (0), low (1), or high (2)
- Remarks:
- The get_hab_parm function called during initialization expects the appropriate habitat-specific parameter to have a first header line that has parameter names that exactly match those in this code.
Definition at line 2463 of file UnitMod.c.
References get_hab_parm(), HP_ALG_MAX, HP_DOM_AEROBTHIN, HP_DOM_MAXDEPTH, HP_FireInt, HP_FLOC_IC, HP_FLOC_IC_CTOOM, HP_FLOC_IC_PC, HP_HYD_POROSITY, HP_HYD_RCINFILT, HP_HYD_SPEC_YIELD, HP_MAC_CANOPDECOUP, HP_MAC_KSP, HP_MAC_LIGHTSAT, HP_MAC_MAXCANOPCOND, HP_MAC_MAXHT, HP_MAC_MAXLAI, HP_MAC_MAXROUGH, HP_MAC_MINROUGH, HP_MAC_SALIN_THRESH, HP_MAC_TEMPOPT, HP_MAC_TRANSLOC_RC, HP_MAC_WAT_TOLER, HP_NPHBIO_IC_CTOOM, HP_NPHBIO_IC_PC, HP_NPHBIO_MAX, HP_NPHBIO_ROOTDEPTH, HP_PHBIO_IC_CTOOM, HP_PHBIO_IC_PC, HP_PHBIO_MAX, HP_PHBIO_RCMORT, HP_PHBIO_RCNPP, HP_PhosHi, HP_PhosInt, HP_PhosLo, HP_SalinHi, HP_SalinInt, HP_SalinLo, HP_SALT_ICSEDWAT, HP_SALT_ICSFWAT, HP_SfDepthHi, HP_SfDepthInt, HP_SfDepthLo, and HP_TP_CONC_GRAD.
02464 { 02465 02466 /* Periphyton/Algae */ 02467 HP_ALG_MAX = get_hab_parm(s_parm_name, s_parm_relval, "HP_ALG_MAX"); 02468 02469 /* Soil */ 02470 HP_DOM_MAXDEPTH = get_hab_parm(s_parm_name, s_parm_relval,"HP_DOM_MAXDEPTH"); 02471 HP_DOM_AEROBTHIN = get_hab_parm(s_parm_name, s_parm_relval,"HP_DOM_AEROBTHIN"); 02472 02473 /* Phosphorus */ 02474 HP_TP_CONC_GRAD = get_hab_parm(s_parm_name, s_parm_relval,"HP_TP_CONC_GRAD"); 02475 02476 /* Salt/tracer */ 02477 HP_SALT_ICSEDWAT = get_hab_parm(s_parm_name, s_parm_relval,"HP_SALT_ICSEDWAT"); 02478 HP_SALT_ICSFWAT = get_hab_parm(s_parm_name, s_parm_relval,"HP_SALT_ICSFWAT"); 02479 02480 /* Macrophytes */ 02481 HP_PHBIO_MAX = get_hab_parm(s_parm_name, s_parm_relval,"HP_PHBIO_MAX"); 02482 HP_NPHBIO_MAX = get_hab_parm(s_parm_name, s_parm_relval,"HP_NPHBIO_MAX"); 02483 HP_MAC_MAXHT = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_MAXHT"); 02484 HP_NPHBIO_ROOTDEPTH = get_hab_parm(s_parm_name, s_parm_relval,"HP_NPHBIO_ROOTDEPTH"); 02485 HP_MAC_MAXROUGH = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_MAXROUGH"); 02486 HP_MAC_MINROUGH = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_MINROUGH"); 02487 HP_MAC_MAXLAI = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_MAXLAI"); 02488 HP_MAC_MAXCANOPCOND = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_MAXCANOPCOND"); /* unused in ELMv2.2, 2.3 */ 02489 HP_MAC_CANOPDECOUP = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_CANOPDECOUP"); /* unused in ELMv2.2, 2.3 */ 02490 HP_MAC_TEMPOPT = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_TEMPOPT"); 02491 HP_MAC_LIGHTSAT = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_LIGHTSAT"); 02492 HP_MAC_KSP = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_KSP"); 02493 HP_PHBIO_RCNPP = get_hab_parm(s_parm_name, s_parm_relval,"HP_PHBIO_RCNPP"); 02494 HP_PHBIO_RCMORT = get_hab_parm(s_parm_name, s_parm_relval,"HP_PHBIO_RCMORT"); 02495 HP_MAC_WAT_TOLER = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_WAT_TOLER"); 02496 HP_MAC_SALIN_THRESH = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_SALIN_THRESH"); /* unused in ELMv2.2, 2.3 */ 02497 HP_PHBIO_IC_CTOOM = get_hab_parm(s_parm_name, s_parm_relval,"HP_PHBIO_IC_CTOOM"); 02498 HP_NPHBIO_IC_CTOOM = get_hab_parm(s_parm_name, s_parm_relval,"HP_NPHBIO_IC_CTOOM"); 02499 HP_PHBIO_IC_PC = get_hab_parm(s_parm_name, s_parm_relval,"HP_PHBIO_IC_PC"); 02500 HP_NPHBIO_IC_PC = get_hab_parm(s_parm_name, s_parm_relval,"HP_NPHBIO_IC_PC"); 02501 HP_MAC_TRANSLOC_RC = get_hab_parm(s_parm_name, s_parm_relval,"HP_MAC_TRANSLOC_RC"); 02502 02503 /* Hydrology */ 02504 HP_HYD_RCINFILT = get_hab_parm(s_parm_name, s_parm_relval,"HP_HYD_RCINFILT"); 02505 HP_HYD_SPEC_YIELD = get_hab_parm(s_parm_name, s_parm_relval,"HP_HYD_SPEC_YIELD"); 02506 HP_HYD_POROSITY = get_hab_parm(s_parm_name, s_parm_relval,"HP_HYD_POROSITY"); 02507 02508 /* Floc */ 02509 HP_FLOC_IC = get_hab_parm(s_parm_name, s_parm_relval,"HP_FLOC_IC"); 02510 HP_FLOC_IC_CTOOM = get_hab_parm(s_parm_name, s_parm_relval,"HP_FLOC_IC_CTOOM"); 02511 HP_FLOC_IC_PC = get_hab_parm(s_parm_name, s_parm_relval,"HP_FLOC_IC_PC"); 02512 02513 /* Habitat succession */ 02514 HP_SfDepthLo = get_hab_parm(s_parm_name, s_parm_relval,"HP_SfDepthLo"); 02515 HP_SfDepthHi = get_hab_parm(s_parm_name, s_parm_relval,"HP_SfDepthHi"); 02516 HP_SfDepthInt = get_hab_parm(s_parm_name, s_parm_relval,"HP_SfDepthInt"); 02517 HP_PhosLo = get_hab_parm(s_parm_name, s_parm_relval,"HP_PhosLo"); 02518 HP_PhosHi = get_hab_parm(s_parm_name, s_parm_relval,"HP_PhosHi"); 02519 HP_PhosInt = get_hab_parm(s_parm_name, s_parm_relval,"HP_PhosInt"); 02520 HP_FireInt = get_hab_parm(s_parm_name, s_parm_relval,"HP_FireInt"); 02521 HP_SalinLo = get_hab_parm(s_parm_name, s_parm_relval,"HP_SalinLo"); 02522 HP_SalinHi = get_hab_parm(s_parm_name, s_parm_relval,"HP_SalinHi"); 02523 HP_SalinInt = get_hab_parm(s_parm_name, s_parm_relval,"HP_SalinInt"); 02524 02525 }

| void ReadModExperimParms | ( | char * | s_parm_name, | |
| int | s_parm_relval | |||
| ) |
Acquire the model parameters that are used in special model experiments - used only in special, research-oriented applications. (Added for v2.6).
NOTE that this was quickly set up for the ELM Peer Review (2006) project, and (at least in v2.6.0) should be used in conjunction with the spatial interpolators for rain and pET inputs. (because this is intended for scales that are potentially incompatible w/ simultaneous use of gridIO inputs on SFWMM stage/depth that are expected w/ gridIO rain/pET).
NOTE: this has not been specifically developed/tested for inclusion within the automated sensitivity analyses - thus only been used at this point for the ModExperimParms_NOM nominal parameter set (Nov 2006).
.
- Parameters:
-
s_parm_name Name of the sensitivity parameter being varied for this run s_parm_relval Indicator of current value of relative range in sensitivity parm values: nominal (0), low (1), or high (2)
Definition at line 2636 of file UnitMod.c.
References conv_kgTOmg, get_modexperim_parm(), GP_BC_InputRow, GP_BC_Pconc, GP_BC_SatWat_add, GP_BC_Sconc, GP_BC_SfWat_add, GP_BC_SfWat_addHi, GP_BC_SfWat_targ, isModExperim, msgStr, usrErr(), usrErr0(), and WriteMsg().
02637 { 02638 if (isModExperim) { 02639 sprintf(msgStr,"####### Reading special model experiment parameters. \nYou should NOT be using gridIO spatial time series data for rain, pET, and SFWMM-output depth #######..."); 02640 WriteMsg(msgStr,1); 02641 usrErr0(msgStr); 02642 02643 GP_BC_SfWat_add = get_modexperim_parm(s_parm_name, s_parm_relval,"GP_BC_SfWat_add"); 02644 GP_BC_SfWat_addHi = get_modexperim_parm(s_parm_name, s_parm_relval,"GP_BC_SfWat_addHi"); 02645 GP_BC_SatWat_add = get_modexperim_parm(s_parm_name, s_parm_relval,"GP_BC_SatWat_add"); 02646 GP_BC_SfWat_targ = get_modexperim_parm(s_parm_name, s_parm_relval,"GP_BC_SfWat_targ"); 02647 GP_BC_InputRow = get_modexperim_parm(s_parm_name, s_parm_relval,"GP_BC_InputRow"); 02648 GP_BC_Pconc = get_modexperim_parm(s_parm_name, s_parm_relval,"GP_BC_Pconc"); /* ug/L */ 02649 GP_BC_Sconc = get_modexperim_parm(s_parm_name, s_parm_relval,"GP_BC_Sconc"); /* g/L */ 02650 02651 GP_BC_Pconc /= conv_kgTOmg; /* convert from ug/L to g/L (same as div by the kg->mg conversion) */ 02652 02653 sprintf(msgStr,"done."); 02654 WriteMsg(msgStr,1); 02655 usrErr(msgStr); 02656 } 02657 else { 02658 GP_BC_SfWat_add = 0.0; /* value of 0.0 has no effect on model results (adds nothing to boundary depth/stages) */ 02659 GP_BC_SfWat_addHi = 0.0; /* value of 0.0 has no effect on model results (adds nothing to boundary depth/stages) */ 02660 GP_BC_SatWat_add = 0.0; /* value of 0.0 has no effect on model results (adds nothing to boundary depth/stages) */ 02661 GP_BC_SfWat_targ = 0.0; /* value of 0.0 has no effect on model results (only used in Oct06 specialized model experiment - code doesn't use this parm otherwise) */ 02662 GP_BC_InputRow = 0; /* value of 0 has no effect on model results (only used in Oct06 specialized model experiment - code doesn't use this parm otherwise) */ 02663 GP_BC_Pconc = 0.0; 02664 GP_BC_Sconc = 0.0; 02665 } 02666 }
| void PtInterp_read | ( | void | ) |
On-the-fly interpolations to develop spatial time series from point data.
.
Read the point data into the lists.
Note that this was quickly set up for the ELM Peer Review (2006) project, and (at least in v2.6.0) needs to be used in conjunction with the model experiment parms for synthetic stage boundary conditions (because it skips all gridIO inputs, including SFWMM stage/depth).
For ELM v2.6, extracted/re-used these code fragments from ELM v2.1 code, with associated updated uses/documentation
Definition at line 2680 of file UnitMod.c.
References NPtTS2, NPtTS3, NPtTS4, NPtTS5, NPtTS6, PTSL_Init(), PTSL_ReadLists(), pTSList2, pTSList3, pTSList4, pTSList5, pTSList6, Timestep2, Timestep3, Timestep4, Timestep5, Timestep6, TIndex2, TIndex3, TIndex4, TIndex5, TIndex6, usrErr(), and usrErr0().
02681 { 02682 /* rainfall and pET point data for models that do not use gridIO inputs, but use interpolated point time series */ 02683 /* final arg in PTSL_ReadLists is the column number of of the data (not incl. the 3 date columns) */ 02684 usrErr0 ("\n*** WARNING: You are not using gridIO spatial time series data for rain, pET, and SFWMM-output depth. \nReading rainfall and pET point time series data... "); 02685 pTSList2 = PTSL_Init(50, 2); 02686 PTSL_ReadLists(pTSList2,"rain",TIndex2++,&Timestep2,&NPtTS2, 1); /* rainfall in data-col 1 (file's col 4) */ 02687 02688 pTSList3 = PTSL_Init(50, 2); 02689 PTSL_ReadLists(pTSList3,"pET",TIndex3++,&Timestep3,&NPtTS3, 1); /* pET data in data-col 1 (file's col 4) */ 02690 02691 usrErr("done.\n"); 02692 02693 if (99 == 66) { /* never-get-here: temporary v2.6 PTSL - Below (old code frags extracted from v2.1) are left here as template for other data (or re-implementing the ELM v2.1 pET calculations if data available) */ 02694 pTSList4 = PTSL_Init(50, 2); 02695 PTSL_ReadLists(pTSList4,"pET",TIndex4++,&Timestep4,&NPtTS4, 2); /* dew point temperature data can be in data-col 2 */ 02696 pTSList5 = PTSL_Init(50, 2); 02697 PTSL_ReadLists(pTSList5,"pET",TIndex5++,&Timestep5,&NPtTS5, 3); /* wind speed data can be in data-col 3 */ 02698 pTSList6 = PTSL_Init(50, 2); 02699 PTSL_ReadLists(pTSList6,"pET",TIndex6++,&Timestep6,&NPtTS6, 4); /* cloud cover data can be in data-col 4 */ 02700 } 02701 }
| void get_map_dims | ( | void | ) |
Get the map dimensions of the global model array.
This mererly calls the "read_map_dims" function
Definition at line 2707 of file UnitMod.c.
References read_map_dims().
02708 { 02709 read_map_dims("Elevation"); 02710 }

| void alloc_memory | ( | ) |
Allocate memory.
Allocate memory for model variables, and initialize (not context-specific) values.
Definition at line 2717 of file UnitMod.c.
References AIR_TEMP, ALG_INCID_LIGHT, ALG_LIGHT_CF, ALG_LIGHT_EXTINCT, ALG_REFUGE, ALG_SAT, ALG_TEMP_CF, ALG_TOT, ALG_WAT_CF, basn, Bathymetry, BCmodel_sfwat, BCondFlow, boundcond_depth, boundcond_ETp, boundcond_rain, BulkD, C_ALG, C_ALG_AVAIL_MORT, C_ALG_GPP, C_ALG_GPP_P, C_ALG_MORT, C_ALG_MORT_P, C_ALG_MORT_POT, C_ALG_NPP, C_ALG_NUT_CF, C_ALG_P, C_ALG_PC, C_ALG_PCrep, C_ALG_PROD_CF, C_ALG_RESP, C_ALG_RESP_POT, DEPOS_ORG_MAT, DIM, DINdummy, DOM_BD, DOM_DECOMP, DOM_DECOMP_POT, DOM_FR_FLOC, DOM_fr_nphBio, DOM_P_OM, DOM_QUALITY_CF, DOM_SED_AEROB_Z, DOM_SED_ANAEROB_Z, DOM_TEMP_CF, DOM_Z, DOP, DOP_DECOMP, DOP_FLOC, DOP_nphBio, ELEVATION, FIREdummy, FLOC, FLOC_DECOMP, FLOC_DECOMP_POT, FLOC_DECOMP_QUAL_CF, FLOC_DEPO, FLOC_DEPO_POT, FLOC_FR_ALGAE, Floc_fr_phBio, FLOC_Z, FlocP, FlocP_DECOMP, FlocP_DEPO, FlocP_FR_ALGAE, FlocP_OM, FlocP_OMrep, FlocP_PhBio, H2O_TEMP, HAB, HP_ALG_MAX, HP_DOM_AEROBTHIN, HP_DOM_MAXDEPTH, HP_FireInt, HP_FLOC_IC, HP_FLOC_IC_CTOOM, HP_FLOC_IC_PC, HP_HYD_POROSITY, HP_HYD_RCINFILT, HP_HYD_SPEC_YIELD, HP_MAC_CANOPDECOUP, HP_MAC_KSP, HP_MAC_LIGHTSAT, HP_MAC_MAXCANOPCOND, HP_MAC_MAXHT, HP_MAC_MAXLAI, HP_MAC_MAXROUGH, HP_MAC_MINROUGH, HP_MAC_SALIN_THRESH, HP_MAC_TEMPOPT, HP_MAC_TRANSLOC_RC, HP_MAC_WAT_TOLER, HP_NPHBIO_IC_CTOOM, HP_NPHBIO_IC_PC, HP_NPHBIO_MAX, HP_NPHBIO_ROOTDEPTH, HP_PHBIO_IC_CTOOM, HP_PHBIO_IC_PC, HP_PHBIO_MAX, HP_PHBIO_RCMORT, HP_PHBIO_RCNPP, HP_PhosHi, HP_PhosInt, HP_PhosLo, HP_SalinHi, HP_SalinInt, HP_SalinLo, HP_SALT_ICSEDWAT, HP_SALT_ICSFWAT, HP_SfDepthHi, HP_SfDepthInt, HP_SfDepthLo, HP_TP_CONC_GRAD, HYD_DOM_ACTWAT_PRES, HYD_DOM_ACTWAT_VOL, HYD_ET, HYD_EVAP_CALC, HYD_MANNINGS_N, HYD_RCCONDUCT, HYD_SAT_POT_TRANS, HYD_SED_WAT_VOL, HYD_TOT_POT_TRANSP, HYD_TRANSP, HYD_UNSAT_POT_TRANS, HYD_WATER_AVAIL, HydRelDepPosNeg, HydTotHd, init_pvar(), Inorg_Z, LAI_eff, MAC_HEIGHT, MAC_LAI, MAC_LIGHT_CF, MAC_MAX_BIO, MAC_NOPH_BIOMAS, mac_nph_CtoOM, mac_nph_OM, mac_nph_P, mac_nph_PC, mac_nph_PC_rep, MAC_NUT_CF, MAC_PH_BIOMAS, mac_ph_CtoOM, mac_ph_OM, mac_ph_P, mac_ph_PC, mac_ph_PC_rep, MAC_PROD_CF, MAC_REL_BIOM, MAC_SALT_CF, MAC_TEMP_CF, MAC_TOT_BIOM, MAC_WATER_AVAIL_CF, MAC_WATER_CF, nalloc(), NC_ALG, NC_ALG_AVAIL_MORT, NC_ALG_GPP, NC_ALG_GPP_P, NC_ALG_MORT, NC_ALG_MORT_P, NC_ALG_MORT_POT, NC_ALG_NPP, NC_ALG_NUT_CF, NC_ALG_P, NC_ALG_PC, NC_ALG_PCrep, NC_ALG_PROD_CF, NC_ALG_RESP, NC_ALG_RESP_POT, NPHBIO_AVAIL, NPHBIO_MORT, nphbio_mort_OM, nphbio_mort_P, NPHBIO_MORT_POT, NPHBIO_REFUGE, NPHBIO_SAT, nphbio_transl_OM, nphbio_transl_P, NPHBIO_TRANSLOC, NPHBIO_TRANSLOC_POT, ON_MAP, P_SUM_CELL, PHBIO_AVAIL, PHBIO_MORT, phbio_mort_OM, phbio_mort_P, PHBIO_MORT_POT, PHBIO_NPP, phbio_npp_OM, phbio_npp_P, PHBIO_REFUGE, PHBIO_SAT, phbio_transl_OM, phbio_transl_P, PHBIO_TRANSLOC, PHBIO_TRANSLOC_POT, s0, s1, SAL_SED_WT, SAL_SF_WT, SAL_SF_WT_mb, SALT_Atm_Depos, SALT_SED_TO_SF_FLOW, SALT_SED_WT, SALT_SFWAT_DOWNFL, SALT_SFWAT_DOWNFL_POT, SALT_SURF_WT, SAT_TO_UNSAT_FL, SAT_VS_UNSAT, SAT_WATER, SAT_WT_HEAD, SAT_WT_RECHG, SAT_WT_TRANSP, SED_ELEV, SED_INACT_Z, SF_WT_EVAP, SF_WT_FROM_RAIN, SF_WT_INFILTRATION, SF_WT_POT_INF, SF_WT_TO_SAT_DOWNFLOW, SF_WT_VEL_mag, SFWT_VOL, soil_MOIST_CF, SOLRAD274, SOLRADGRD, SURFACE_WAT, TP_Act_to_Tot, TP_Act_to_TotRep, TP_Atm_Depos, TP_AtmosDepos, TP_DNFLOW, TP_DNFLOW_POT, TP_K, TP_SED_CONC, TP_SED_MINER, TP_SED_WT, TP_SED_WT_AZ, TP_SEDWT_CONCACT, TP_SEDWT_CONCACTMG, TP_SEDWT_UPTAKE, TP_settl, TP_settlDays, TP_SF_WT, TP_SFWT_CONC, TP_SFWT_CONC_MG, TP_SFWT_MINER, TP_SFWT_UPTAK, TP_SORB, TP_SORB_POT, TP_SORBCONC, TP_SORBCONC_rep, TP_SORBTION, TP_UPFLOW, TP_UPFLOW_POT, TPtoSOIL, TPtoSOIL_rep, TPtoVOL, TPtoVOL_rep, UNSAT_AVAIL, UNSAT_CAP, UNSAT_DEPTH, UNSAT_HYD_COND_CF, UNSAT_MOIST_PRP, UNSAT_PERC, UNSAT_TO_SAT_FL, UNSAT_TRANSP, UNSAT_WATER, UNSAT_WT_POT, usrErr(), usrErr0(), and WQMsettlVel.
02718 { 02719 usrErr0("Allocating Memory..."); /* console message */ 02720 02721 ON_MAP = (unsigned char *) nalloc(sizeof(unsigned char)*(s0+2)*(s1+2),"ON_MAP"); 02722 init_pvar(ON_MAP,NULL,'c',0.0); 02723 02724 BCondFlow = (int *) nalloc(sizeof(int)*(s0+2)*(s1+2),"BCondFlow"); 02725 init_pvar(BCondFlow,NULL,'d',0.0); 02726 HAB = (unsigned char *) nalloc(sizeof(unsigned char)*(s0+2)*(s1+2),"HAB"); 02727 init_pvar(HAB,NULL,'c',0.0); 02728 basn = (int *) nalloc(sizeof(int)*(s0+2)*(s1+2),"basn"); /* Basin/Indicator-Region map */ 02729 init_pvar(basn,NULL,'d',0.0); 02730 02731 ALG_INCID_LIGHT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_INCID_LIGHT"); 02732 init_pvar(ALG_INCID_LIGHT,NULL,'f',0.0); 02733 ALG_LIGHT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_LIGHT_CF"); 02734 init_pvar(ALG_LIGHT_CF,NULL,'f',0.0); 02735 ALG_LIGHT_EXTINCT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_LIGHT_EXTINCT"); 02736 init_pvar(ALG_LIGHT_EXTINCT,NULL,'f',0.0); 02737 ALG_WAT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_WAT_CF"); 02738 init_pvar(ALG_WAT_CF,NULL,'f',0.0); 02739 ALG_TEMP_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_TEMP_CF"); 02740 init_pvar(ALG_TEMP_CF,NULL,'f',0.0); 02741 ALG_REFUGE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_REFUGE"); 02742 init_pvar(ALG_REFUGE,NULL,'f',0.0); 02743 ALG_SAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_SAT"); 02744 init_pvar(ALG_SAT,NULL,'f',0.0); 02745 ALG_TOT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ALG_TOT"); 02746 init_pvar(ALG_TOT,NULL,'f',0.0); 02747 02748 NC_ALG_AVAIL_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_AVAIL_MORT"); 02749 init_pvar(NC_ALG_AVAIL_MORT,NULL,'f',0.0); 02750 NC_ALG_GPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_GPP"); 02751 init_pvar(NC_ALG_GPP,NULL,'f',0.0); 02752 NC_ALG_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_MORT"); 02753 init_pvar(NC_ALG_MORT,NULL,'f',0.0); 02754 NC_ALG_MORT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_MORT_POT"); 02755 init_pvar(NC_ALG_MORT_POT,NULL,'f',0.0); 02756 NC_ALG_NPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_NPP"); 02757 init_pvar(NC_ALG_NPP,NULL,'f',0.0); 02758 NC_ALG_NUT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_NUT_CF"); 02759 init_pvar(NC_ALG_NUT_CF,NULL,'f',0.0); 02760 NC_ALG_PROD_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_PROD_CF"); 02761 init_pvar(NC_ALG_PROD_CF,NULL,'f',0.0); 02762 NC_ALG_RESP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_RESP"); 02763 init_pvar(NC_ALG_RESP,NULL,'f',0.0); 02764 NC_ALG_RESP_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_RESP_POT"); 02765 init_pvar(NC_ALG_RESP_POT,NULL,'f',0.0); 02766 NC_ALG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG"); 02767 init_pvar(NC_ALG,NULL,'f',0.0); 02768 C_ALG_AVAIL_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_AVAIL_MORT"); 02769 init_pvar(C_ALG_AVAIL_MORT,NULL,'f',0.0); 02770 C_ALG_GPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_GPP"); 02771 init_pvar(C_ALG_GPP,NULL,'f',0.0); 02772 C_ALG_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_MORT"); 02773 init_pvar(C_ALG_MORT,NULL,'f',0.0); 02774 C_ALG_MORT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_MORT_POT"); 02775 init_pvar(C_ALG_MORT_POT,NULL,'f',0.0); 02776 C_ALG_NPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_NPP"); 02777 init_pvar(C_ALG_NPP,NULL,'f',0.0); 02778 C_ALG_NUT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_NUT_CF"); 02779 init_pvar(C_ALG_NUT_CF,NULL,'f',0.0); 02780 C_ALG_PROD_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_PROD_CF"); 02781 init_pvar(C_ALG_PROD_CF,NULL,'f',0.0); 02782 C_ALG_RESP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_RESP"); 02783 init_pvar(C_ALG_RESP,NULL,'f',0.0); 02784 C_ALG_RESP_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_RESP_POT"); 02785 init_pvar(C_ALG_RESP_POT,NULL,'f',0.0); 02786 C_ALG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG"); 02787 init_pvar(C_ALG,NULL,'f',0.0); 02788 NC_ALG_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_P"); 02789 init_pvar(NC_ALG_P,NULL,'f',0.0); 02790 C_ALG_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_P"); 02791 init_pvar(C_ALG_P,NULL,'f',0.0); 02792 NC_ALG_GPP_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_GPP_P"); 02793 init_pvar(NC_ALG_GPP_P,NULL,'f',0.0); 02794 C_ALG_GPP_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_GPP_P"); 02795 init_pvar(C_ALG_GPP_P,NULL,'f',0.0); 02796 NC_ALG_MORT_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_MORT_P"); 02797 init_pvar(NC_ALG_MORT_P,NULL,'f',0.0); 02798 C_ALG_MORT_P = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_MORT_P"); 02799 init_pvar(C_ALG_MORT_P,NULL,'f',0.0); 02800 NC_ALG_PCrep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_ALG_PCrep"); 02801 init_pvar(NC_ALG_PCrep,NULL,'f',0.0); 02802 C_ALG_PCrep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_ALG_PCrep"); 02803 init_pvar(C_ALG_PCrep,NULL,'f',0.0); 02804 NC_ALG_PC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"NC_ALG_PC"); 02805 init_pvar(NC_ALG_PC,NULL,'l',0.0); 02806 C_ALG_PC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"C_ALG_PC"); 02807 init_pvar(C_ALG_PC,NULL,'l',0.0); 02808 02809 DEPOS_ORG_MAT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"DEPOS_ORG_MAT"); 02810 init_pvar(DEPOS_ORG_MAT,NULL,'l',0.0); 02811 02812 DOM_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_Z"); 02813 init_pvar(DOM_Z,NULL,'f',0.0); 02814 DOM_DECOMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_DECOMP"); 02815 init_pvar(DOM_DECOMP,NULL,'f',0.0); 02816 DOM_DECOMP_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_DECOMP_POT"); 02817 init_pvar(DOM_DECOMP_POT,NULL,'f',0.0); 02818 DOM_fr_nphBio = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_fr_nphBio"); 02819 init_pvar(DOM_fr_nphBio,NULL,'f',0.0); 02820 02821 Floc_fr_phBio = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Floc_fr_phBio"); 02822 init_pvar(Floc_fr_phBio,NULL,'f',0.0); 02823 DOM_FR_FLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_FR_FLOC"); 02824 init_pvar(DOM_FR_FLOC,NULL,'f',0.0); 02825 soil_MOIST_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"soil_MOIST_CF"); 02826 init_pvar(soil_MOIST_CF,NULL,'f',0.0); 02827 DOM_QUALITY_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_QUALITY_CF"); 02828 init_pvar(DOM_QUALITY_CF,NULL,'f',0.0); 02829 DOM_SED_AEROB_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_SED_AEROB_Z"); 02830 init_pvar(DOM_SED_AEROB_Z,NULL,'f',0.0); 02831 DOM_SED_ANAEROB_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_SED_ANAEROB_Z"); 02832 init_pvar(DOM_SED_ANAEROB_Z,NULL,'f',0.0); 02833 DOM_TEMP_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_TEMP_CF"); 02834 init_pvar(DOM_TEMP_CF,NULL,'f',0.0); 02835 ELEVATION = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ELEVATION"); 02836 init_pvar(ELEVATION,NULL,'f',0.0); 02837 Bathymetry = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Bathymetry"); 02838 init_pvar(Bathymetry,NULL,'f',0.0); 02839 SED_ELEV = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SED_ELEV"); 02840 init_pvar(SED_ELEV,NULL,'f',0.0); 02841 SED_INACT_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SED_INACT_Z"); 02842 init_pvar(SED_INACT_Z,NULL,'f',0.0); 02843 DOP_FLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOP_FLOC"); 02844 init_pvar(DOP_FLOC,NULL,'f',0.0); 02845 DOP_nphBio = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOP_nphBio"); 02846 init_pvar(DOP_nphBio,NULL,'f',0.0); 02847 02848 DOM_P_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"DOM_P_OM"); 02849 init_pvar(DOM_P_OM,NULL,'l',0.0); 02850 02851 TPtoSOIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoSOIL"); 02852 init_pvar(TPtoSOIL,NULL,'f',0.0); 02853 TPtoSOIL_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoSOIL_rep"); 02854 init_pvar(TPtoSOIL_rep,NULL,'f',0.0); 02855 TPtoVOL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoVOL"); 02856 init_pvar(TPtoVOL,NULL,'f',0.0); 02857 TPtoVOL_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoVOL_rep"); 02858 init_pvar(TPtoVOL_rep,NULL,'f',0.0); 02859 BulkD = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"BulkD"); 02860 init_pvar(BulkD,NULL,'f',0.0); 02861 DOM_BD = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOM_BD"); 02862 init_pvar(DOM_BD,NULL,'f',0.0); 02863 Inorg_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Inorg_Z"); 02864 init_pvar(Inorg_Z,NULL,'f',0.0); 02865 DIM = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DIM"); 02866 init_pvar(DIM,NULL,'f',0.0); 02867 DOP_DECOMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DOP_DECOMP"); 02868 init_pvar(DOP_DECOMP,NULL,'f',0.0); 02869 02870 DOP = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"DOP"); 02871 init_pvar(DOP,NULL,'l',0.0); 02872 02873 /* placeholder for fire */ 02874 FIREdummy = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FIREdummy"); 02875 init_pvar(FIREdummy,NULL,'f',0.0); 02876 02877 HYD_DOM_ACTWAT_PRES = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_DOM_ACTWAT_PRES"); 02878 init_pvar(HYD_DOM_ACTWAT_PRES,NULL,'f',0.0); 02879 HYD_DOM_ACTWAT_VOL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_DOM_ACTWAT_VOL"); 02880 init_pvar(HYD_DOM_ACTWAT_VOL,NULL,'f',0.0); 02881 HYD_EVAP_CALC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_EVAP_CALC"); 02882 init_pvar(HYD_EVAP_CALC,NULL,'f',0.0); 02883 HYD_MANNINGS_N = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_MANNINGS_N"); 02884 init_pvar(HYD_MANNINGS_N,NULL,'f',0.0); 02885 HYD_RCCONDUCT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_RCCONDUCT"); 02886 init_pvar(HYD_RCCONDUCT,NULL,'f',0.0); 02887 HYD_SAT_POT_TRANS = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_SAT_POT_TRANS"); 02888 init_pvar(HYD_SAT_POT_TRANS,NULL,'f',0.0); 02889 HYD_SED_WAT_VOL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_SED_WAT_VOL"); 02890 init_pvar(HYD_SED_WAT_VOL,NULL,'f',0.0); 02891 HYD_TOT_POT_TRANSP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_TOT_POT_TRANSP"); 02892 init_pvar(HYD_TOT_POT_TRANSP,NULL,'f',0.0); 02893 HydTotHd = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HydTotHd"); 02894 init_pvar(HydTotHd,NULL,'f',0.0); 02895 HydRelDepPosNeg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HydRelDepPosNeg"); 02896 init_pvar(HydRelDepPosNeg,NULL,'f',0.0); 02897 HYD_TRANSP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_TRANSP"); 02898 init_pvar(HYD_TRANSP,NULL,'f',0.0); 02899 HYD_ET = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_ET"); 02900 init_pvar(HYD_ET,NULL,'f',0.0); 02901 HYD_UNSAT_POT_TRANS = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_UNSAT_POT_TRANS"); 02902 init_pvar(HYD_UNSAT_POT_TRANS,NULL,'f',0.0); 02903 HYD_WATER_AVAIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HYD_WATER_AVAIL"); 02904 init_pvar(HYD_WATER_AVAIL,NULL,'f',0.0); 02905 02906 /* sfwmm rainfall, stage, and pET mapped to elm grid */ 02907 boundcond_rain = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"boundcond_rain"); 02908 init_pvar(boundcond_rain,NULL,'f',0.0); 02909 boundcond_depth = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"boundcond_depth"); 02910 init_pvar(boundcond_depth,NULL,'f',0.0); 02911 boundcond_ETp = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"boundcond_ETp"); 02912 init_pvar(boundcond_ETp,NULL,'f',0.0); 02913 02914 SAT_TO_UNSAT_FL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_TO_UNSAT_FL"); 02915 init_pvar(SAT_TO_UNSAT_FL,NULL,'f',0.0); 02916 SAT_VS_UNSAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_VS_UNSAT"); 02917 init_pvar(SAT_VS_UNSAT,NULL,'f',0.0); 02918 SAT_WATER = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_WATER"); 02919 init_pvar(SAT_WATER,NULL,'f',0.0); 02920 SAT_WT_HEAD = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_WT_HEAD"); 02921 init_pvar(SAT_WT_HEAD,NULL,'f',0.0); 02922 SAT_WT_RECHG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_WT_RECHG"); 02923 init_pvar(SAT_WT_RECHG,NULL,'f',0.0); 02924 SAT_WT_TRANSP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAT_WT_TRANSP"); 02925 init_pvar(SAT_WT_TRANSP,NULL,'f',0.0); 02926 SF_WT_EVAP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_EVAP"); 02927 init_pvar(SF_WT_EVAP,NULL,'f',0.0); 02928 SF_WT_FROM_RAIN = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_FROM_RAIN"); 02929 init_pvar(SF_WT_FROM_RAIN,NULL,'f',0.0); 02930 SF_WT_INFILTRATION = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_INFILTRATION"); 02931 init_pvar(SF_WT_INFILTRATION,NULL,'f',0.0); 02932 SF_WT_POT_INF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_POT_INF"); 02933 init_pvar(SF_WT_POT_INF,NULL,'f',0.0); 02934 SF_WT_TO_SAT_DOWNFLOW = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_TO_SAT_DOWNFLOW"); 02935 init_pvar(SF_WT_TO_SAT_DOWNFLOW,NULL,'f',0.0); 02936 SFWT_VOL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SFWT_VOL"); 02937 init_pvar(SFWT_VOL,NULL,'f',0.0); 02938 SURFACE_WAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SURFACE_WAT"); 02939 init_pvar(SURFACE_WAT,NULL,'f',0.0); 02940 SF_WT_VEL_mag = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_VEL_mag"); 02941 init_pvar(SF_WT_VEL_mag,NULL,'f',0.0); 02942 BCmodel_sfwat = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"BCmodel_sfwat"); 02943 init_pvar(BCmodel_sfwat,NULL,'f',0.0); 02944 02945 UNSAT_AVAIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_AVAIL"); 02946 init_pvar(UNSAT_AVAIL,NULL,'f',0.0); 02947 UNSAT_CAP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_CAP"); 02948 init_pvar(UNSAT_CAP,NULL,'f',0.0); 02949 UNSAT_DEPTH = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_DEPTH"); 02950 init_pvar(UNSAT_DEPTH,NULL,'f',0.0); 02951 UNSAT_HYD_COND_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_HYD_COND_CF"); 02952 init_pvar(UNSAT_HYD_COND_CF,NULL,'f',0.0); 02953 UNSAT_MOIST_PRP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_MOIST_PRP"); 02954 init_pvar(UNSAT_MOIST_PRP,NULL,'f',0.0); 02955 UNSAT_PERC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_PERC"); 02956 init_pvar(UNSAT_PERC,NULL,'f',0.0); 02957 UNSAT_TO_SAT_FL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_TO_SAT_FL"); 02958 init_pvar(UNSAT_TO_SAT_FL,NULL,'f',0.0); 02959 UNSAT_TRANSP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_TRANSP"); 02960 init_pvar(UNSAT_TRANSP,NULL,'f',0.0); 02961 UNSAT_WATER = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_WATER"); 02962 init_pvar(UNSAT_WATER,NULL,'f',0.0); 02963 UNSAT_WT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UNSAT_WT_POT"); 02964 init_pvar(UNSAT_WT_POT,NULL,'f',0.0); 02965 02966 MAC_HEIGHT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_HEIGHT"); 02967 init_pvar(MAC_HEIGHT,NULL,'f',0.0); 02968 MAC_LAI = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_LAI"); 02969 init_pvar(MAC_LAI,NULL,'f',0.0); 02970 LAI_eff = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"LAI_eff"); 02971 init_pvar(LAI_eff,NULL,'f',0.0); 02972 MAC_LIGHT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_LIGHT_CF"); 02973 init_pvar(MAC_LIGHT_CF,NULL,'f',0.0); 02974 MAC_MAX_BIO = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_MAX_BIO"); 02975 init_pvar(MAC_MAX_BIO,NULL,'f',0.0); 02976 MAC_NOPH_BIOMAS = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_NOPH_BIOMAS"); 02977 init_pvar(MAC_NOPH_BIOMAS,NULL,'f',0.0); 02978 MAC_NUT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_NUT_CF"); 02979 init_pvar(MAC_NUT_CF,NULL,'f',0.0); 02980 MAC_PH_BIOMAS = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_PH_BIOMAS"); 02981 init_pvar(MAC_PH_BIOMAS,NULL,'f',0.0); 02982 MAC_PROD_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_PROD_CF"); 02983 init_pvar(MAC_PROD_CF,NULL,'f',0.0); 02984 MAC_REL_BIOM = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_REL_BIOM"); 02985 init_pvar(MAC_REL_BIOM,NULL,'f',0.0); 02986 MAC_SALT_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_SALT_CF"); 02987 init_pvar(MAC_SALT_CF,NULL,'f',0.0); 02988 MAC_TEMP_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_TEMP_CF"); 02989 init_pvar(MAC_TEMP_CF,NULL,'f',0.0); 02990 MAC_TOT_BIOM = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_TOT_BIOM"); 02991 init_pvar(MAC_TOT_BIOM,NULL,'f',0.0); 02992 MAC_WATER_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_WATER_CF"); 02993 init_pvar(MAC_WATER_CF,NULL,'f',0.0); 02994 MAC_WATER_AVAIL_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MAC_WATER_AVAIL_CF"); 02995 init_pvar(MAC_WATER_AVAIL_CF,NULL,'f',0.0); 02996 NPHBIO_AVAIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_AVAIL"); 02997 init_pvar(NPHBIO_AVAIL,NULL,'f',0.0); 02998 NPHBIO_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_MORT"); 02999 init_pvar(NPHBIO_MORT,NULL,'f',0.0); 03000 NPHBIO_MORT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_MORT_POT"); 03001 init_pvar(NPHBIO_MORT_POT,NULL,'f',0.0); 03002 NPHBIO_REFUGE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_REFUGE"); 03003 init_pvar(NPHBIO_REFUGE,NULL,'f',0.0); 03004 NPHBIO_SAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_SAT"); 03005 init_pvar(NPHBIO_SAT,NULL,'f',0.0); 03006 NPHBIO_TRANSLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_TRANSLOC"); 03007 init_pvar(NPHBIO_TRANSLOC,NULL,'f',0.0); 03008 NPHBIO_TRANSLOC_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NPHBIO_TRANSLOC_POT"); 03009 init_pvar(NPHBIO_TRANSLOC_POT,NULL,'f',0.0); 03010 PHBIO_AVAIL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_AVAIL"); 03011 init_pvar(PHBIO_AVAIL,NULL,'f',0.0); 03012 PHBIO_MORT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_MORT"); 03013 init_pvar(PHBIO_MORT,NULL,'f',0.0); 03014 PHBIO_MORT_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_MORT_POT"); 03015 init_pvar(PHBIO_MORT_POT,NULL,'f',0.0); 03016 PHBIO_NPP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_NPP"); 03017 init_pvar(PHBIO_NPP,NULL,'f',0.0); 03018 PHBIO_REFUGE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_REFUGE"); 03019 init_pvar(PHBIO_REFUGE,NULL,'f',0.0); 03020 PHBIO_SAT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_SAT"); 03021 init_pvar(PHBIO_SAT,NULL,'f',0.0); 03022 PHBIO_TRANSLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_TRANSLOC"); 03023 init_pvar(PHBIO_TRANSLOC,NULL,'f',0.0); 03024 PHBIO_TRANSLOC_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PHBIO_TRANSLOC_POT"); 03025 init_pvar(PHBIO_TRANSLOC_POT,NULL,'f',0.0); 03026 03027 phbio_npp_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_npp_P"); 03028 init_pvar(phbio_npp_P,NULL,'l',0.0); 03029 phbio_npp_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_npp_OM"); 03030 init_pvar(phbio_npp_OM,NULL,'l',0.0); 03031 phbio_mort_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_mort_P"); 03032 init_pvar(phbio_mort_P,NULL,'l',0.0); 03033 phbio_mort_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_mort_OM"); 03034 init_pvar(phbio_mort_OM,NULL,'l',0.0); 03035 phbio_transl_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_transl_P"); 03036 init_pvar(phbio_transl_P,NULL,'l',0.0); 03037 phbio_transl_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"phbio_transl_OM"); 03038 init_pvar(phbio_transl_OM,NULL,'l',0.0); 03039 nphbio_transl_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"nphbio_transl_P"); 03040 init_pvar(nphbio_transl_P,NULL,'l',0.0); 03041 nphbio_transl_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"nphbio_transl_OM"); 03042 init_pvar(nphbio_transl_OM,NULL,'l',0.0); 03043 nphbio_mort_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"nphbio_mort_P"); 03044 init_pvar(nphbio_mort_P,NULL,'l',0.0); 03045 nphbio_mort_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"nphbio_mort_OM"); 03046 init_pvar(nphbio_mort_OM,NULL,'l',0.0); 03047 mac_nph_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_nph_P"); 03048 init_pvar(mac_nph_P,NULL,'l',0.0); 03049 mac_nph_PC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_nph_PC"); 03050 init_pvar(mac_nph_PC,NULL,'l',0.0); 03051 mac_nph_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_nph_OM"); 03052 init_pvar(mac_nph_OM,NULL,'l',0.0); 03053 mac_nph_CtoOM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_nph_CtoOM"); 03054 init_pvar(mac_nph_CtoOM,NULL,'l',0.0); 03055 mac_ph_P = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_ph_P"); 03056 init_pvar(mac_ph_P,NULL,'l',0.0); 03057 mac_ph_PC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_ph_PC"); 03058 init_pvar(mac_ph_PC,NULL,'l',0.0); 03059 mac_ph_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_ph_OM"); 03060 init_pvar(mac_ph_OM,NULL,'l',0.0); 03061 mac_ph_CtoOM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"mac_ph_CtoOM"); 03062 init_pvar(mac_ph_CtoOM,NULL,'l',0.0); 03063 03064 mac_nph_PC_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"mac_nph_PC_rep"); 03065 init_pvar(mac_nph_PC_rep,NULL,'f',0.0); 03066 mac_ph_PC_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"mac_ph_PC_rep"); 03067 init_pvar(mac_ph_PC_rep,NULL,'f',0.0); 03068 03069 TP_SED_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SED_WT"); 03070 init_pvar(TP_SED_WT,NULL,'l',0.0); 03071 TP_SED_CONC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SED_CONC"); 03072 init_pvar(TP_SED_CONC,NULL,'l',0.0); 03073 TP_SEDWT_CONCACT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SEDWT_CONCACT"); 03074 init_pvar(TP_SEDWT_CONCACT,NULL,'l',0.0); 03075 TP_SF_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SF_WT"); 03076 init_pvar(TP_SF_WT,NULL,'l',0.0); 03077 TP_SFWT_CONC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SFWT_CONC"); 03078 init_pvar(TP_SFWT_CONC,NULL,'l',0.0); 03079 TP_SORB = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SORB"); 03080 init_pvar(TP_SORB,NULL,'l',0.0); 03081 TP_SORBCONC = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SORBCONC"); 03082 init_pvar(TP_SORBCONC,NULL,'l',0.0); 03083 TP_SED_WT_AZ = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_SED_WT_AZ"); 03084 init_pvar(TP_SED_WT_AZ,NULL,'l',0.0); 03085 TP_Act_to_Tot = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"TP_Act_to_Tot"); 03086 init_pvar(TP_Act_to_Tot,NULL,'l',0.0); 03087 03088 TP_Act_to_TotRep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_Act_to_TotRep"); 03089 init_pvar(TP_Act_to_TotRep,NULL,'f',0.0); 03090 TP_SORBCONC_rep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SORBCONC_rep"); 03091 init_pvar(TP_SORBCONC_rep,NULL,'f',0.0); 03092 03093 TP_DNFLOW = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_DNFLOW"); 03094 init_pvar(TP_DNFLOW,NULL,'f',0.0); 03095 TP_DNFLOW_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_DNFLOW_POT"); 03096 init_pvar(TP_DNFLOW_POT,NULL,'f',0.0); 03097 TP_Atm_Depos = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_Atm_Depos"); 03098 init_pvar(TP_Atm_Depos,NULL,'f',0.0); 03099 /* v2.6 added map of atmospheric TP deposition rate */ 03100 TP_AtmosDepos = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_AtmosDepos"); 03101 init_pvar(TP_AtmosDepos,NULL,'f',0.0); 03102 TP_K = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_K"); 03103 init_pvar(TP_K,NULL,'f',0.0); 03104 TP_SED_MINER = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SED_MINER"); 03105 init_pvar(TP_SED_MINER,NULL,'f',0.0); 03106 TP_SEDWT_CONCACTMG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SEDWT_CONCACTMG"); 03107 init_pvar(TP_SEDWT_CONCACTMG,NULL,'f',0.0); 03108 TP_SEDWT_UPTAKE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SEDWT_UPTAKE"); 03109 init_pvar(TP_SEDWT_UPTAKE,NULL,'f',0.0); 03110 TP_SFWT_CONC_MG = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SFWT_CONC_MG"); 03111 init_pvar(TP_SFWT_CONC_MG,NULL,'f',0.0); 03112 TP_SFWT_MINER = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SFWT_MINER"); 03113 init_pvar(TP_SFWT_MINER,NULL,'f',0.0); 03114 TP_SFWT_UPTAK = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SFWT_UPTAK"); 03115 init_pvar(TP_SFWT_UPTAK,NULL,'f',0.0); 03116 TP_settl = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_settl"); 03117 init_pvar(TP_settl,NULL,'f',0.0); 03118 TP_SORB_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SORB_POT"); 03119 init_pvar(TP_SORB_POT,NULL,'f',0.0); 03120 TP_SORBTION = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_SORBTION"); 03121 init_pvar(TP_SORBTION,NULL,'f',0.0); 03122 TP_UPFLOW = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_UPFLOW"); 03123 init_pvar(TP_UPFLOW,NULL,'f',0.0); 03124 TP_UPFLOW_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_UPFLOW_POT"); 03125 init_pvar(TP_UPFLOW_POT,NULL,'f',0.0); 03126 P_SUM_CELL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"P_SUM_CELL"); 03127 init_pvar(P_SUM_CELL,NULL,'f',0.0); 03128 03129 DINdummy = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"DINdummy"); 03130 init_pvar(DINdummy,NULL,'l',0.0); 03131 03132 WQMsettlVel = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"WQMsettlVel"); 03133 init_pvar(WQMsettlVel,NULL,'f',0.0); 03134 TP_settlDays = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_settlDays"); 03135 init_pvar(TP_settlDays,NULL,'f',0.0); 03136 03137 SAL_SED_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"SAL_SED_WT"); 03138 init_pvar(SAL_SED_WT,NULL,'l',0.0); 03139 SAL_SF_WT_mb = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"SAL_SF_WT_mb"); 03140 init_pvar(SAL_SF_WT_mb,NULL,'l',0.0); 03141 SALT_SED_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"SALT_SED_WT"); 03142 init_pvar(SALT_SED_WT,NULL,'l',0.0); 03143 SALT_SURF_WT = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"SALT_SURF_WT"); 03144 init_pvar(SALT_SURF_WT,NULL,'l',0.0); 03145 03146 SAL_SF_WT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SAL_SF_WT"); 03147 init_pvar(SAL_SF_WT,NULL,'f',0.0); 03148 SALT_Atm_Depos = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SALT_Atm_Depos"); 03149 init_pvar(SALT_Atm_Depos,NULL,'f',0.0); 03150 SALT_SED_TO_SF_FLOW = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SALT_SED_TO_SF_FLOW"); 03151 init_pvar(SALT_SED_TO_SF_FLOW,NULL,'f',0.0); 03152 SALT_SFWAT_DOWNFL = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SALT_SFWAT_DOWNFL"); 03153 init_pvar(SALT_SFWAT_DOWNFL,NULL,'f',0.0); 03154 SALT_SFWAT_DOWNFL_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SALT_SFWAT_DOWNFL_POT"); 03155 init_pvar(SALT_SFWAT_DOWNFL_POT,NULL,'f',0.0); 03156 03157 FLOC_DECOMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DECOMP"); 03158 init_pvar(FLOC_DECOMP,NULL,'f',0.0); 03159 FLOC_DECOMP_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DECOMP_POT"); 03160 init_pvar(FLOC_DECOMP_POT,NULL,'f',0.0); 03161 FLOC_DECOMP_QUAL_CF = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DECOMP_QUAL_CF"); 03162 init_pvar(FLOC_DECOMP_QUAL_CF,NULL,'f',0.0); 03163 FLOC_Z = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_Z"); 03164 init_pvar(FLOC_Z,NULL,'f',0.0); 03165 FLOC_DEPO = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DEPO"); 03166 init_pvar(FLOC_DEPO,NULL,'f',0.0); 03167 FLOC_DEPO_POT = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_DEPO_POT"); 03168 init_pvar(FLOC_DEPO_POT,NULL,'f',0.0); 03169 FLOC_FR_ALGAE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC_FR_ALGAE"); 03170 init_pvar(FLOC_FR_ALGAE,NULL,'f',0.0); 03171 FLOC = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FLOC"); 03172 init_pvar(FLOC,NULL,'f',0.0); 03173 FlocP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP"); 03174 init_pvar(FlocP,NULL,'f',0.0); 03175 FlocP_DECOMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_DECOMP"); 03176 init_pvar(FlocP_DECOMP,NULL,'f',0.0); 03177 FlocP_FR_ALGAE = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_FR_ALGAE"); 03178 init_pvar(FlocP_FR_ALGAE,NULL,'f',0.0); 03179 FlocP_PhBio = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_PhBio"); 03180 init_pvar(FlocP_PhBio,NULL,'f',0.0); 03181 FlocP_DEPO = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_DEPO"); 03182 init_pvar(FlocP_DEPO,NULL,'f',0.0); 03183 FlocP_OMrep = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"FlocP_OMrep"); 03184 init_pvar(FlocP_OMrep,NULL,'f',0.0); 03185 FlocP_OM = (double *) nalloc(sizeof(double)*(s0+2)*(s1+2),"FlocP_OM"); 03186 init_pvar(FlocP_OM,NULL,'l',0.0); 03187 03188 SOLRADGRD = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SOLRADGRD"); 03189 init_pvar(SOLRADGRD,NULL,'f',0.0); 03190 SOLRAD274 = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SOLRAD274"); 03191 init_pvar(SOLRAD274,NULL,'f',0.0); 03192 AIR_TEMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"AIR_TEMP"); 03193 init_pvar(AIR_TEMP,NULL,'f',0.0); 03194 H2O_TEMP = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"H2O_TEMP"); 03195 init_pvar(H2O_TEMP,NULL,'f',0.0); 03196 03197 /* habitat-specific parameter arrays */ 03198 HP_ALG_MAX = NULL; 03199 03200 HP_DOM_MAXDEPTH = NULL; 03201 HP_DOM_AEROBTHIN = NULL; 03202 03203 HP_TP_CONC_GRAD = NULL; 03204 03205 HP_SALT_ICSEDWAT = NULL; 03206 HP_SALT_ICSFWAT = NULL; 03207 03208 HP_PHBIO_MAX = NULL; 03209 HP_NPHBIO_MAX = NULL; 03210 HP_MAC_MAXHT = NULL; 03211 HP_NPHBIO_ROOTDEPTH = NULL; 03212 HP_MAC_MAXROUGH = NULL; 03213 HP_MAC_MINROUGH = NULL; 03214 HP_MAC_MAXLAI = NULL; 03215 HP_MAC_MAXCANOPCOND = NULL; 03216 HP_MAC_CANOPDECOUP = NULL; 03217 HP_MAC_TEMPOPT = NULL; 03218 HP_MAC_LIGHTSAT = NULL; 03219 HP_MAC_KSP = NULL; 03220 HP_PHBIO_RCNPP = NULL; 03221 HP_PHBIO_RCMORT = NULL; 03222 HP_MAC_WAT_TOLER = NULL; 03223 HP_MAC_SALIN_THRESH = NULL; 03224 HP_PHBIO_IC_CTOOM = NULL; 03225 HP_NPHBIO_IC_CTOOM = NULL; 03226 HP_PHBIO_IC_PC = NULL; 03227 HP_NPHBIO_IC_PC = NULL; 03228 HP_MAC_TRANSLOC_RC = NULL; 03229 03230 HP_HYD_RCINFILT = NULL; 03231 HP_HYD_SPEC_YIELD = NULL; 03232 HP_HYD_POROSITY = NULL; 03233 03234 HP_FLOC_IC = NULL; 03235 HP_FLOC_IC_CTOOM = NULL; 03236 HP_FLOC_IC_PC = NULL; 03237 03238 HP_SfDepthLo = NULL; 03239 HP_SfDepthHi = NULL; 03240 HP_SfDepthInt = NULL; 03241 HP_PhosLo = NULL; 03242 HP_PhosHi = NULL; 03243 HP_PhosInt = NULL; 03244 HP_FireInt = NULL; 03245 HP_SalinLo = NULL; 03246 HP_SalinHi = NULL; 03247 HP_SalinInt = NULL; 03248 03249 usrErr("Done."); /* console message */ 03250 03251 }
| float graph7 | ( | unsigned char | y, | |
| float | x | |||
| ) |
Time series interplator, data set 7.
The graphX functions are generic time-series interpolator; graph7 is interpolator for data set g7 (determining unsaturated infiltration control function).
- Parameters:
-
y NA (=0) x independent variable data value (soil moisture proportion)
- Returns:
- float dependent variable value (proportion of max infiltration rate)
- Variables local to function
s local slope of independent/dependent variable values
ig array index of the data pairs
Np number of data pairs (incl. 0)
Definition at line 3265 of file UnitMod.c.
References g7.
Referenced by cell_dyn7().
03266 { 03267 float s; 03268 int ig=0, Np=10; 03275 while(1) { 03276 if (x <= g7[ig][0]) { if(ig>0) ig=ig-1; else return(g7[0][1+y]);} 03277 else if (x > g7[ig][0] && x <= g7[ig+1][0]) { 03278 s = (g7[ig+1][1+y]-g7[ig][1+y])/ 03279 (g7[ig+1][0]-g7[ig][0]); 03280 return(s * (x-g7[ig][0]) + g7[ig][1+y]); } 03281 else if (x > g7[ig+1][0]) { if(ig<Np) ig=ig+1; else return(g7[Np][1+y]);} 03282 } 03283 }
| float graph8 | ( | unsigned char | y, | |
| float | x | |||
| ) |
Time series interplator, data set 8.
The graphX functions are generic time-series interpolator; graph8 is interpolator for data set g8 (determining water availability to plants).
- Parameters:
-
y NA (=0) x independent variable data value (proportion of water available in upper soil zone)
- Returns:
- float dependent variable value (proportion of max water availability to plants)
- Varialbes local to function
s local slope of independent/dependent variable values
ig array index of the data pairs
Np number of data pairs (incl. 0)
Definition at line 3292 of file UnitMod.c.
References g8.
Referenced by cell_dyn7(), and init_eqns().
03293 { 03294 float s; 03295 int ig=0, Np=10; 03302 while(1) { 03303 if (x <= g8[ig][0]) { if(ig>0) ig=ig-1; else return(g8[0][1+y]);} 03304 else if (x > g8[ig][0] && x <= g8[ig+1][0]) { 03305 s = (g8[ig+1][1+y]-g8[ig][1+y])/ 03306 (g8[ig+1][0]-g8[ig][0]); 03307 return(s * (x-g8[ig][0]) + g8[ig][1+y]); } 03308 else if (x > g8[ig+1][0]) { if(ig<Np) ig=ig+1; else return(g8[Np][1+y]);} 03309 } 03310 }
| void HabSwitch_Init | ( | void | ) |
Transfers habitat-specific parameters into data struct, allocate memory.
Definition at line 25 of file Success.c.
Referenced by init_succession().
00026 { 00027 int ii; 00028 00029 /* put these habitat-specific parameters into a concise data struct for use here */ 00030 for( ii = 0; ii < habNumTot; ii++) { 00031 Habi[ii].Water.Llo = HP_SfDepthLo[ii+1]; 00032 Habi[ii].Water.Lhi = HP_SfDepthHi[ii+1]; 00033 Habi[ii].Water.Pin = HP_SfDepthInt[ii+1]; 00034 Habi[ii].Nutrient.Llo = HP_PhosLo[ii+1]; 00035 Habi[ii].Nutrient.Lhi = HP_PhosHi[ii+1]; 00036 Habi[ii].Nutrient.Pin = HP_PhosInt[ii+1]; 00037 Habi[ii].PFin = HP_FireInt[ii+1]; 00038 Habi[ii].Salinity.Llo = HP_SalinLo[ii+1]; 00039 Habi[ii].Salinity.Lhi = HP_SalinHi[ii+1]; 00040 Habi[ii].Salinity.Pin = HP_SalinInt[ii+1]; 00041 } 00042 00043 alloc_hab_hist( ); 00044 sprintf (msgStr, "Succession ON, module OK for %d habitats...", ii ); usrErr(msgStr); WriteMsg(msgStr,1); 00045 00046 return; 00047 }
| unsigned char HabSwitch | ( | int | ix, | |
| int | iy, | |||
| float * | Water, | |||
| float * | Nutrient, | |||
| int * | Fire, | |||
| float * | Salinity, | |||
| unsigned char * | HAB | |||
| ) |
Switches habitats (the option that is currently used).
Returns the number of habitat to switch to based on the length of the weekly (or other) averaged period of favorable conditions. In this case the period of habitat favorable conditions is calculated as the number of weeks (or other time intervals) during which the habitat conditions matched the specified for a period longer than the given one - SW_TIME_TH. The switching occurs as soon as the number of such successfull weeks exceeds the time specified in Pin
- Parameters:
-
ix Model domain row iy Model domain column Water Current water depth data array Nutrient Current nutrient conc. data array Fire Fire data array (unused, no fire implemented) Salinity Current salinity conc. data array HAB Habitat-type data array
- Returns:
- habitat type of cell
Definition at line 66 of file Success.c.
Referenced by cell_dyn1().
00067 { 00068 int StateW, StateN, StateS, i; 00069 int HabW[MAX_NHAB], HabN[MAX_NHAB], HabS[MAX_NHAB], DW, DN, DS; 00070 int cell = T(ix,iy); 00071 int hab = HAB[cell]; /* current habitat in the cell */ 00072 00073 /* define the habitat type that matches the existing water conditions */ 00074 00075 /* HabHist is an array of integers : ssSnnNwwW, where 00076 ww are the two digits for the number of weeks in the water level favorable conditions, 00077 W the number of days in those conditions during the current week; 00078 nn are the two digits for the number of weeks being in the nutrient favorable conditions, 00079 N the number of days in those conditions during the current week; 00080 ss are the two digits for the number of weeks in the salinity favorable conditions, 00081 S the number of days in those conditions during the current week; 00082 00083 The switching occurs when all of these histories exceed the habitat specific 00084 periods Pin. 00085 */ 00086 for ( i = 0; i < habNumTot; i++ ) 00087 { 00088 DW = HabHist[cell*habNumTot +i]%10; 00089 HabW[i] = (HabHist[cell*habNumTot +i]/10)%100; 00090 DN = (HabHist[cell*habNumTot +i]/1000)%10; 00091 HabN[i] = HabHist[cell*habNumTot +i]/10000; 00092 DS = (HabHist[cell*habNumTot +i]/1000000)%10; 00093 HabS[i] = HabHist[cell*habNumTot +i]/10000000; 00094 00095 /* when the averaging time elapses, if #favorable days exceeds a threshold (using 4 for 7 day AV_PER), increment the period (weeks) history */ 00096 if ((int)SimTime.TIME%AV_PER == 0) 00097 { 00098 if (DW > SW_TIME_TH_W) HabHist[cell*habNumTot +i] = HabHist[cell*habNumTot +i] - DW + 100; 00099 else HabHist[cell*habNumTot +i] = 0; 00100 if (DN > SW_TIME_TH_N) HabHist[cell*habNumTot +i] = HabHist[cell*habNumTot +i] - DN*1000 + 10000; 00101 else HabHist[cell*habNumTot +i] = 0; 00102 if (DS > SW_TIME_TH_S) HabHist[cell*habNumTot +i] = HabHist[cell*habNumTot +i] - DS*1000000 + 10000000; 00103 else HabHist[cell*habNumTot +i] = 0; 00104 } 00105 00106 /* check what habitat type the existing conditions match; increment the day# if conditions are favorable */ 00107 if ( InHab (Water[cell], Habi[i].Water) ) HabHist[cell*habNumTot + i] = HabHist[cell*habNumTot + i] + 1; 00108 if ( InHab (Nutrient[cell]*conv_kgTOmg, Habi[i].Nutrient) ) HabHist[cell*habNumTot + i] = HabHist[cell*habNumTot + i] + 1000; 00109 if ( InHab (Salinity[cell], Habi[i].Salinity) ) HabHist[cell*habNumTot + i] = HabHist[cell*habNumTot + i] + 1000000; 00110 } 00111 00112 /* check if the historical conditions for switching hold */ 00113 for ( i = 0; i < habNumTot; i++ ) 00114 if ( HabW[i] >= Habi[i].Water.Pin && HabN[i] >= Habi[i].Nutrient.Pin && HabS[i] >= Habi[i].Salinity.Pin ) 00115 { HabHist[cell*habNumTot +i] = 0; 00116 return (i+1); /* returns new hab, HAB[] is offset by 1 from these array IDs */ 00117 } 00118 00119 return hab; 00120 }
| void Run_Canal_Network | ( | float * | SWH, | |
| float * | Elevation, | |||
| float * | MC, | |||
| float * | GW, | |||
| float * | poros, | |||
| float * | GWcond, | |||
| double * | NA, | |||
| double * | PA, | |||
| double * | SA, | |||
| double * | GNA, | |||
| double * | GPA, | |||
| double * | GSA, | |||
| float * | Unsat, | |||
| float * | sp_yield | |||
| ) |
Runs the water management network.
Invoke the calls to water control structure flows and the canal-cell interactions.
- Parameters:
-
SWH SURFACE_WAT Elevation SED_ELEV MC HYD_MANNINGS_N GW SAT_WATER poros HP_HYD_POROSITY GWcond HYD_RCCONDUCT NA DINdummy PA TP_SF_WT SA SALT_SURF_WT GNA DINdummy GPA TP_SED_WT GSA SALT_SED_WT Unsat UNSAT_WATER sp_yield HP_HYD_SPEC_YIELD
Definition at line 330 of file WatMgmt.c.
References Chan::area, Chan::basin, CanalOutFile, CanalOutFile_P, CanalOutFile_S, canDebugFile, canPrint, canstep, Chan_list, simTime::da, debug, Flows_in_Structures(), FluxChannel(), FMOD(), hyd_iter, simTime::IsBudgEnd, simTime::IsSimulationEnd, simTime::mo, N_c_iter, num_chan, P, Chan::P_con, printchan, Chan::S_con, SensiOn, SimTime, Chan::sumHistIn, Chan::sumHistOut, Chan::sumRuleIn, Chan::sumRuleOut, TOT_P_CAN, TOT_S_CAN, TOT_VOL_CAN, Chan::wat_depth, WstructOutFile, WstructOutFile_P, WstructOutFile_S, and simTime::yr.
00334 { int i; 00335 float CH_vol; 00336 00337 if ( canPrint && (( N_c_iter++ % hyd_iter ) == 0.0) ) { 00338 if (SensiOn) { 00339 printchan = (SimTime.IsSimulationEnd) ? (1) : (0); /* flag to indicate to print canal/struct data */ 00340 } 00341 else printchan = 1; 00342 } 00343 else printchan = 0; 00344 00345 if (printchan == 1) { 00346 00347 fprintf( WstructOutFile, "\n%d/%d/%d\t",SimTime.yr[0],SimTime.mo[0],SimTime.da[0] ); 00348 fprintf( WstructOutFile_P, "\n%d/%d/%d\t",SimTime.yr[0],SimTime.mo[0],SimTime.da[0] ); 00349 /* fprintf( WstructOutFile_N, "\n%d/%d/%d\t",SimTime.yr[0],SimTime.mo[0],SimTime.da[0] ); */ 00350 fprintf( WstructOutFile_S, "\n%d/%d/%d\t",SimTime.yr[0],SimTime.mo[0],SimTime.da[0] ); 00351 00352 fprintf (CanalOutFile, "\n%d/%d/%d\t",SimTime.yr[0],SimTime.mo[0],SimTime.da[0] ); 00353 fprintf (CanalOutFile_P, "\n%d/%d/%d\t",SimTime.yr[0],SimTime.mo[0],SimTime.da[0] ); 00354 /* fprintf (CanalOutFile_N, "\n%d/%d/%d\t",SimTime.yr[0],SimTime.mo[0],SimTime.da[0] ); */ 00355 fprintf (CanalOutFile_S, "\n%d/%d/%d\t",SimTime.yr[0],SimTime.mo[0],SimTime.da[0] ); 00356 } 00357 00358 00359 for ( i = 0; i < num_chan; i++ ) 00360 { /* set the sum of historical (or other-model) and Rule-based flows to/from a canal per iteration to 0 */ 00361 Chan_list[i]->sumHistIn = Chan_list[i]->sumHistOut = 0.0; 00362 Chan_list[i]->sumRuleIn = Chan_list[i]->sumRuleOut = 0.0; 00363 } 00364 00365 /* calculate flows through water control structures */ 00366 Flows_in_Structures ( SWH, GW, poros, Elevation, NA, PA, SA ); 00367 00368 /* output header for canal-cell flux output canal debug file, at relatively high debug levels (writes a LOT of stuff) */ 00369 if( debug > 3 ) 00370 { 00371 fprintf( canDebugFile, "N%3d WatDepth Pconc Sconc flux_S flux_G flux_L Qin Qout #_iter Area error \n", N_c_iter ); 00372 00373 } 00374 00375 /* Flux the water along a canal using the relaxation procedure */ 00376 00377 for( i = 0; i < num_chan; i++ ) 00378 if (Chan_list[i]->width > 0) /* a neg width canal is skipped */ 00379 00380 { 00381 FluxChannel ( i, SWH, Elevation, MC, GW, poros, GWcond, NA, PA, SA, GNA, GPA, GSA, Unsat, sp_yield ); 00382 /* calculate total canal volume after canal fluxes */ 00383 CH_vol = Chan_list[i]->area*Chan_list[i]->wat_depth; 00384 00385 /* sum the storages on iteration that performs budget checks */ 00386 if ( !( FMOD(N_c_iter,(1/canstep) )) && (SimTime.IsBudgEnd ) ) { 00387 TOT_VOL_CAN[Chan_list[i]->basin] += CH_vol; 00388 TOT_VOL_CAN[0] += CH_vol; 00389 TOT_P_CAN[Chan_list[i]->basin] += Chan_list[i]->P; 00390 TOT_P_CAN[0] += Chan_list[i]->P; 00391 TOT_S_CAN[Chan_list[i]->basin] += Chan_list[i]->S; 00392 TOT_S_CAN[0] += Chan_list[i]->S; 00393 } 00394 00395 00396 if ( printchan ) 00397 { 00398 /* report N&P concentration in mg/L (kg/m3==g/L * 1000 = mg/L) */ 00399 Chan_list[i]->P_con = (CH_vol > 0) ? (Chan_list[i]->P/CH_vol*1000.0) : 0.0; 00400 /* Chan_list[i]->N_con = (CH_vol > 0) ? (Chan_list[i]->N/CH_vol*1000.0) : 0.0; */ 00401 Chan_list[i]->S_con = (CH_vol > 0) ? (Chan_list[i]->S/CH_vol ) : 0.0; 00402 00403 /* v2.2 increased decimal places in output (from 6.3f to 7.4f) */ 00404 fprintf( CanalOutFile, "%6.2f\t", Chan_list[i]->wat_depth ); 00405 fprintf( CanalOutFile_P, "%7.4f\t", Chan_list[i]->P_con ); 00406 /* fprintf( CanalOutFile_N, "%7.4f\t", Chan_list[i]->N_con ); */ 00407 fprintf( CanalOutFile_S, "%7.4f\t", Chan_list[i]->S_con ); 00408 } 00409 } 00410 if ( printchan ) 00411 { 00412 fflush(CanalOutFile); 00413 fflush(CanalOutFile_P); 00414 /* fflush(CanalOutFile_N); */ 00415 fflush(CanalOutFile_S); 00416 } 00417 00418 return; 00419 }
| void Canal_Network_Init | ( | float | baseDatum, | |
| float * | elev | |||
| ) |
Initialize the water managment network topology.
Initialize/configure the network of canals/levees and and water control structures. The canal information stored in the global array Chan_list
- Parameters:
-
baseDatum GP_DATUM_DISTANCE elev SED_ELEV
Definition at line 96 of file WatMgmt.c.
References Chan::area, CanalOutFile, CanalOutFile_P, CanalOutFile_S, canDebugFile, Chan_list, ChanInFile, Channel_configure(), debug, H_OPSYS, Chan::ic_depth, Chan::ic_N_con, Chan::ic_P_con, Chan::ic_S_con, init_pvar(), MCopen, modelFileName, modelName, modelVers, Chan::N, nalloc(), Structure::next_in_list, num_chan, ON_MAP, OutputPath, Chan::P, ProjName, ReadChanStruct(), ReadStructures(), Chan::S, s0, s1, Structure::S_nam, SimAlt, SimModif, struct_first, structs, T, UNIX, usrErr(), WstructOutFile, WstructOutFile_P, and WstructOutFile_S.
00098 { struct Structure *structs; 00099 int i,j; 00100 char filename[30]; 00101 00102 sprintf(filename,"CanalData"); 00103 00104 /* Input the canals geometry file, return pointer to first vector */ 00105 /* Configure the canals, marking the cells that interact with them */ 00106 /* and storing the pointers to the cells for each of the canals in Chan_list array */ 00107 Channel_configure(elev, ReadChanStruct (filename)); 00108 00109 for( i = 0; i < num_chan; i++ ) 00110 { 00111 Chan_list[i]->P = Chan_list[i]->ic_P_con * Chan_list[i]->area * Chan_list[i]->ic_depth; /* kg/m3 * m2 * m */ 00112 Chan_list[i]->N = Chan_list[i]->ic_N_con * Chan_list[i]->area * Chan_list[i]->ic_depth; /* kg/m3 * m2 * m */ 00113 Chan_list[i]->S = Chan_list[i]->ic_S_con * Chan_list[i]->area * Chan_list[i]->ic_depth; /* kg/m3 * m2 * m */ 00114 } 00115 00116 usrErr ("\tCanal geometry configured OK"); 00117 00118 /* Read data on the water control structures, return pointer to the first structure */ 00119 ReadStructures (filename, baseDatum); 00120 00121 ON_MAP[T(2,2)] = 0; 00122 00123 MCopen = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MCopen"); /* open-water manning's coef, used in structure cell-cell bridge flow */ 00124 init_pvar(MCopen,NULL,'f',0.05); /* this is a hard-wired parameter (value set to last argument, 0.05) - but do not do anything much with it now for the bridge flows */ 00125 00126 00127 if ( debug > 0 ) /* Output the canal/levee modifications to ON_MAP */ 00128 { 00129 sprintf( modelFileName, "%s/%s/Output/Debug/ON_MAP_CANAL.txt", OutputPath, ProjName ); 00130 00131 00132 if ( ( ChanInFile = fopen( modelFileName, "w" ) ) == NULL ) 00133 { 00134 printf( "Can't open the %s canal/levee debug file!\n", modelFileName ); 00135 exit(-1) ; 00136 } 00137 fprintf ( ChanInFile, "ROWS=%d\nCOLUMNS=%d\nFORMAT=text\nINFO=\"Debug data: Overland flow restrictions due to levees. \"\nMISSING=0\n", s0, s1 ); 00138 00139 for ( i = 1; i <= s0 ; i++ ) 00140 { for ( j = 1 ; j <= s1 ; j++ ) 00141 fprintf( ChanInFile, "%4d\t", ON_MAP[T(i,j)]) ; 00142 fprintf ( ChanInFile, "\n" ); 00143 } 00144 fclose( ChanInFile ) ; 00145 } 00146 00147 00148 /* Open file for writing structure flows */ 00149 { if(H_OPSYS==UNIX) 00150 sprintf( modelFileName, "%s/%s/Output/Canal/structsOut", OutputPath, ProjName ); 00151 else sprintf( modelFileName, "%s%s:Output:structsOut", OutputPath, ProjName ); 00152 } 00153 00154 /* Open file to print structure flows to */ 00155 if ( ( WstructOutFile = fopen( modelFileName, "w" ) ) == NULL ) 00156 { 00157 printf( "Can't open %s file!\n", modelFileName ); 00158 exit(-1) ; 00159 } 00160 /* Print The Header Line For This File */ 00161 fprintf ( WstructOutFile, 00162 "%s %s %s %s scenario: Sum of water flows at structures (ft^3/sec)\n Date\t", &modelName, &modelVers, &SimAlt, &SimModif ); 00163 structs = struct_first; 00164 while ( structs != NULL ) 00165 { fprintf( WstructOutFile, "%7s\t", structs->S_nam); 00166 structs = structs->next_in_list; 00167 } 00168 00169 /* Open file for writing structure P concs */ 00170 { if(H_OPSYS==UNIX) 00171 sprintf( modelFileName, "%s/%s/Output/Canal/structsOut_P", OutputPath, ProjName ); 00172 else sprintf( modelFileName, "%s%s:Output:structsOut_P", OutputPath, ProjName ); 00173 } 00174 00175 /* Open file to print structure flows to */ 00176 if ( ( WstructOutFile_P = fopen( modelFileName, "w" ) ) == NULL ) 00177 { 00178 printf( "Can't open %s file!\n", modelFileName ); 00179 exit(-1) ; 00180 } 00181 /* Print The Header Line For This File */ 00182 fprintf ( WstructOutFile_P, 00183 "%s %s %s %s scenario: Flow weighted mean TP conc at structures ( mg/L) \n Date\t", &modelName, &modelVers, &SimAlt, &SimModif ); 00184 structs = struct_first; 00185 while ( structs != NULL ) 00186 { fprintf( WstructOutFile_P, "%7s\t", structs->S_nam); 00187 structs = structs->next_in_list; 00188 } 00189 00190 /* Open file for writing structure Salin concs */ 00191 { if(H_OPSYS==UNIX) 00192 sprintf( modelFileName, "%s/%s/Output/Canal/structsOut_S", OutputPath, ProjName ); 00193 else sprintf( modelFileName, "%s%s:Output:structsOut_S", OutputPath, ProjName ); 00194 } 00195 00196 /* Open file to print structure flows to */ 00197 if ( ( WstructOutFile_S = fopen( modelFileName, "w" ) ) == NULL ) 00198 { 00199 printf( "Can't open %s file!\n", modelFileName ); 00200 exit(-1) ; 00201 } 00202 /* Print The Header Line For This File */ 00203 fprintf ( WstructOutFile_S, 00204 "%s %s %s %s scenario: Flow weighted mean tracer concentration at structures ( g/L) \n Date\t", &modelName, &modelVers, &SimAlt, &SimModif ); 00205 structs = struct_first; 00206 while ( structs != NULL ) 00207 { fprintf( WstructOutFile_S, "%7s\t", structs->S_nam); 00208 structs = structs->next_in_list; 00209 } 00210 00211 /****/ 00212 00213 /* Open file to print canal stage info to */ 00214 { if(H_OPSYS==UNIX) 00215 sprintf( modelFileName, "%s/%s/Output/Canal/CanalOut", OutputPath, ProjName ); 00216 else sprintf( modelFileName, "%s%s:Output:CanalOut", OutputPath, ProjName ); 00217 } 00218 00219 if ( ( CanalOutFile = fopen( modelFileName, "w" ) ) == NULL ) 00220 { 00221 printf( "Can't open %s file!\n", modelFileName ); 00222 exit(-1) ; 00223 } 00224 00225 fprintf ( CanalOutFile, "%s %s %s %s scenario: Instantaneous water depth (meters, from bottom of canal) in canal Reaches\n Date\t", &modelName, &modelVers, &SimAlt, &SimModif ); /* print the header line for this file */ 00226 /* channels with negative widths are reserved for new canals */ 00227 /* v2.2 put a "R_" prefix in front of the canal ID# */ 00228 for( i = 0; i < num_chan; i++ ) {if (Chan_list[i]->width > 0) fprintf ( CanalOutFile, "R_%d\t",Chan_list[i]->number);} 00229 00230 00231 /* Open file to print canal phosph info to */ 00232 { if(H_OPSYS==UNIX) 00233 sprintf( modelFileName, "%s/%s/Output/Canal/CanalOut_P", OutputPath, ProjName ); 00234 else sprintf( modelFileName, "%s%s:Output:CanalOut_P", OutputPath, ProjName ); 00235 } 00236 00237 if ( ( CanalOutFile_P = fopen( modelFileName, "w" ) ) == NULL ) 00238 { 00239 printf( "Can't open %s file!\n", modelFileName ); 00240 exit(-1) ; 00241 } 00242 00243 fprintf ( CanalOutFile_P, "%s %s %s %s scenario: Instantaneous TP conc (mg/L) in canal Reaches\n Date\t", &modelName, &modelVers, &SimAlt, &SimModif ); /* print the header line for this file */ 00244 /* v2.2 put a "R_" prefix in front of the canal ID# */ 00245 for( i = 0; i < num_chan; i++ ) {if (Chan_list[i]->width > 0) fprintf ( CanalOutFile_P, "R_%d\t",Chan_list[i]->number);} 00246 00247 /* Open file to print canal salinity/tracer info to */ 00248 { if(H_OPSYS==UNIX) 00249 sprintf( modelFileName, "%s/%s/Output/Canal/CanalOut_S", OutputPath, ProjName ); 00250 else sprintf( modelFileName, "%s%s:Output:CanalOut_S", OutputPath, ProjName ); 00251 } 00252 00253 if ( ( CanalOutFile_S = fopen( modelFileName, "w" ) ) == NULL ) 00254 { 00255 printf( "Can't open %s file!\n", modelFileName ); 00256 exit(-1) ; 00257 } 00258 00259 fprintf ( CanalOutFile_S, "%s %s %s %s scenario: Instantaneous tracer conc (g/L) in canal Reaches\n Date\t", &modelName, &modelVers, &SimAlt, &SimModif ); /* print the header line for this file */ 00260 /* v2.2 put a "R_" prefix in front of the canal ID# */ 00261 for( i = 0; i < num_chan; i++ ) {if (Chan_list[i]->width > 0) fprintf ( CanalOutFile_S, "R_%d\t",Chan_list[i]->number);} 00262 00263 00264 00265 /* Open file for canal-cell flux debugging purposes */ 00266 if( debug > 0 ) 00267 { 00268 if(H_OPSYS==UNIX) 00269 sprintf( modelFileName, "%s/%s/Output/Debug/Can_debug", OutputPath, ProjName ); 00270 else sprintf( modelFileName, "%s%s:Output:Can_debug", OutputPath, ProjName ); 00271 00272 00273 /* Open file for debugging and error/warnings on canal-cell flows, structure flows */ 00274 if ( ( canDebugFile = fopen( modelFileName, "w" ) ) == NULL ) 00275 { 00276 printf( "Can't open %s file!\n", modelFileName ); 00277 exit(-1) ; 00278 } 00279 00280 fprintf ( canDebugFile, "Depending on level of the debug requested, printed here are data on canal-cell flows and excessive demands on data-driven structure flows. \n" ); /* v2.5 made use of this file pointer */ 00281 fflush(canDebugFile); 00282 00283 } 00284 00285 return; 00286 00287 }

| void CanalReInit | ( | ) |
Re-initialize the depths and concentrations in canals under the Positional Analysis mode.
Definition at line 292 of file WatMgmt.c.
References Chan::area, Chan_list, Chan::ic_depth, Chan::ic_N_con, Chan::ic_P_con, Chan::ic_S_con, Chan::N, num_chan, Chan::P, Chan::S, and Chan::wat_depth.
00293 { 00294 int i; 00295 for( i = 0; i < num_chan; i++ ) 00296 00297 { 00298 Chan_list[i]->wat_depth = Chan_list[i]->ic_depth; 00299 Chan_list[i]->P = Chan_list[i]->ic_P_con * Chan_list[i]->area * Chan_list[i]->ic_depth; /* kg/m3 * m2 * m */ 00300 Chan_list[i]->N = Chan_list[i]->ic_N_con * Chan_list[i]->area * Chan_list[i]->ic_depth; /* kg/m3 * m2 * m */ 00301 Chan_list[i]->S = Chan_list[i]->ic_S_con * Chan_list[i]->area * Chan_list[i]->ic_depth; /* kg/m3 * m2 * m */ 00302 00303 } 00304 }
| void getInt | ( | FILE * | inFile, | |
| const char * | lString, | |||
| int * | iValPtr | |||
| ) |
Get an integer following a specific string.
- Parameters:
-
inFile File that is open lString The string being read iValPtr The char value found
Definition at line 1740 of file Driver_Utilities.c.
References scan_forward().
01741 { 01742 int test; 01743 UCHAR iEx=0; 01744 if(lString) 01745 scan_forward(inFile,lString); 01746 test = fscanf(inFile,"%d",iValPtr); 01747 if(test != 1) { 01748 fprintf(stderr,"Read Error (%d):",test); 01749 if(lString) fprintf(stderr,"%s\n",lString); 01750 *iValPtr = 0; 01751 iEx = 1; 01752 } 01753 if (iEx) exit(0); 01754 }

| void read_map_dims | ( | const char * | filename | ) |
Establish the number of rows and columns of the model.
Read the map dimensions of the global model array, establishing rows=s0 and columns=s1
- Parameters:
-
filename character string of the representative map (usually "Elevation")
Definition at line 975 of file Driver_Utilities.c.
References broadcastInt(), debug, Exit(), gbl_size, Lprocnum, modelFileName, ModelPath, msgStr, ProjName, s0, s1, scan_forward(), usrErr(), and WriteMsg().
00976 { 00977 FILE *file; 00978 00979 if (debug>3) { sprintf(msgStr,"Getting map dims: %s",filename); usrErr(msgStr);} 00980 if(Lprocnum == 1) { 00981 sprintf(modelFileName,"%s/%s/Data/Map_head/%s",ModelPath,ProjName,filename); 00982 file = fopen(modelFileName,"r"); 00983 if(file==NULL) { 00984 fprintf(stderr,"Unable to open map header file %s.\n",modelFileName); 00985 fflush(stderr); 00986 Exit(0); 00987 } 00988 scan_forward(file,"cols:"); 00989 fscanf(file,"%d",&gbl_size[1]); 00990 sprintf(msgStr,"cols = %d\n",gbl_size[1]); WriteMsg(msgStr,1); 00991 scan_forward(file,"rows:"); 00992 fscanf(file,"%d",&gbl_size[0]); 00993 sprintf(msgStr,"rows = %d\n",gbl_size[0]); WriteMsg(msgStr,1); 00994 fclose(file); 00995 } 00996 broadcastInt(gbl_size); /* does nothing in serial (non-parallel) implementation */ 00997 broadcastInt(gbl_size+1); /* does nothing in serial (non-parallel) implementation */ 00998 s0 = gbl_size[0]; 00999 s1 = gbl_size[1]; 01000 }
| void write_output | ( | int | index, | |
| ViewParm * | vp, | |||
| void * | Map, | |||
| const char * | filename, | |||
| char | Mtype, | |||
| int | step | |||
| ) |
Determine which functions to call for model output.
- Parameters:
-
index Count index of the output iteration for this variable (may be > step) vp A struct of ViewParm, outlist configuration Map Model variable array data filename Output file name Mtype General output format type step The current output step
Definition at line 435 of file Driver_Utilities.c.
References calc_maxmin(), debug, viewParm::fileName, point3D::format, getPrecision(), viewParm::mapType, msgStr, viewParm::nPoints, scale1::o, viewParm::points, print_loc_ave(), print_point(), quick_look(), scale1::s, viewParm::scale, viewParm::step, point3D::type, viewParm::varUnits, write_map_file(), WriteMsg(), point3D::x, point3D::y, and point3D::z.
00437 { 00438 int i; Point3D point; 00439 00440 00441 if(vp->mapType != 'N' ) { 00442 write_map_file((char*)filename,Map,Mtype,index,vp->scale.s,vp->scale.o,getPrecision(vp),vp->mapType,vp->varUnits,vp->step,vp->fileName); /* v2.8.2 added varUnits, vp->step (which is outstep interval), and fileName for netCDF use */ 00443 if (debug > 3) calc_maxmin(vp,Map,Mtype,(char*)filename,step); 00444 } 00445 for(i=0; i< (vp->nPoints); i++ ) { 00446 point = vp->points[i]; 00447 if (debug > 3) { sprintf(msgStr,"\nwriting Out: %s(%d): %c(), index = %d, step=%d\n", filename, i, point.type, index, step ); 00448 WriteMsg(msgStr,1); 00449 } 00450 00451 switch( point.type ) { 00452 case 'm': case 'M': calc_maxmin( vp,Map,Mtype,(char*)filename,step); break; 00453 case 'w': quick_look( Map,(char*) filename, Mtype, point.x, point.y, point.z, point.format ); break; 00454 case 'a': case 's': case 'A': case 'S': print_loc_ave( &point, Map, Mtype, (char*)filename, step ); break; 00455 case 'p': print_point( vp, Map, Mtype, (char*)filename, step, i ); break; 00456 } 00457 } 00458 }
| void getChar | ( | FILE * | inFile, | |
| const char * | lString, | |||
| char * | cValPtr | |||
| ) |
Get a character following a specific string.
- Parameters:
-
inFile File that is open lString The string being read cValPtr The char value found
Definition at line 1702 of file Driver_Utilities.c.
References scan_forward().
01703 { 01704 int test; 01705 UCHAR iEx=0; 01706 if(lString) scan_forward(inFile,lString); 01707 test = fscanf(inFile,"%c",cValPtr); 01708 if(test != 1) { 01709 fprintf(stderr,"Read Error (%d):",test); 01710 if(lString) fprintf(stderr,"%s\n",lString); 01711 iEx = 1; 01712 *cValPtr = '\0'; 01713 } 01714 if (iEx) exit(0); 01715 }

| void init_pvar | ( | VOIDP | Map, | |
| UCHAR * | mask, | |||
| unsigned char | Mtype, | |||
| float | iv | |||
| ) |
Initialize a variable to a value.
- Parameters:
-
Map array of data mask data mask Mtype the data type of the map data iv the value used to initialize variable
Definition at line 1008 of file Driver_Utilities.c.
01009 { 01010 int i0, i1; 01011 01012 switch(Mtype) { 01013 case 'b' : /* added double for (non-map) basin (b) array budget calcs */ 01014 for(i0=0; i0<=numBasn; i0++) { 01015 ((double*)Map)[i0] = iv; 01016 } 01017 break; 01018 case 'l' : /* added double (l == letter "ell" ) for map arrays */ 01019 for(i0=0; i0<=s0+1; i0++) 01020 for(i1=0; i1<=s1+1; i1++) { 01021 if(mask==NULL) ((double*)Map)[T(i0,i1)] = iv; 01022 else if ( mask[T(i0,i1)] == 0 ) ((double*)Map)[T(i0,i1)] = 0; 01023 else ((double*)Map)[T(i0,i1)] = iv; 01024 } 01025 break; 01026 case 'f' : 01027 for(i0=0; i0<=s0+1; i0++) 01028 for(i1=0; i1<=s1+1; i1++) { 01029 if(mask==NULL) ((float*)Map)[T(i0,i1)] = iv; 01030 else if ( mask[T(i0,i1)] == 0 ) ((float*)Map)[T(i0,i1)] = 0; 01031 else ((float*)Map)[T(i0,i1)] = iv; 01032 } 01033 break; 01034 case 'i' : case 'd' : 01035 for(i0=0; i0<=s0+1; i0++) 01036 for(i1=0; i1<=s1+1; i1++) { 01037 if(mask==NULL) ((int*)Map)[T(i0,i1)] = (int)iv; 01038 else if ( mask[T(i0,i1)] == 0 ) ((int*)Map)[T(i0,i1)] = 0; 01039 else ((int*)Map)[T(i0,i1)] = (int)iv; 01040 } 01041 break; 01042 case 'c' : 01043 for(i0=0; i0<=s0+1; i0++) 01044 for(i1=0; i1<=s1+1; i1++) { 01045 if(mask==NULL) ((unsigned char*)Map)[T(i0,i1)] = (unsigned char) '\0' + (int) iv; 01046 else if ( mask[T(i0,i1)] == 0 ) ((unsigned char*)Map)[T(i0,i1)] = '\0'; 01047 else ((unsigned char*)Map)[T(i0,i1)] = (unsigned char) '\0' + (int) iv; 01048 } 01049 break; 01050 01051 default : 01052 printf(" in default case\n"); 01053 break; 01054 } 01055 }
| int read_map_file | ( | const char * | filename, | |
| VOIDP | Map, | |||
| unsigned char | Mtype, | |||
| float | scale_value, | |||
| float | offset_value | |||
| ) |
Get the file and read/convert the data of map (array).
- Parameters:
-
filename Input file name Map Model variable array data Mtype General data format type scale_value Variable-scaling multiplier offset_value Variable-scaling offset
- Returns:
- byte size of data values
Definition at line 674 of file Driver_Utilities.c.
References dbgPt, debug, gridSize, gTemp, gTempSize, lcl_size, link_edges(), msgStr, nalloc(), procnum, quick_look(), readMap(), s0, s1, T, usrErr(), WriteMsg(), point2D::x, and point2D::y.
00677 { 00678 int i, j, k0, size; 00679 unsigned temp; 00680 UCHAR *tmpPtr, Dsize; 00681 00682 gridSize = (s0+2)*(s1+2); 00683 gTempSize = gridSize*8; 00684 00685 00686 size = gridSize*4 +1; 00687 if( size > gTempSize ) { 00688 if(gTemp) free((char*)gTemp); 00689 gTemp = (UCHAR*)nalloc( size, "gTemp" ); 00690 gTempSize = size; 00691 } 00692 00693 if(debug>2) { 00694 sprintf(msgStr,"Reading %s\n",filename); 00695 usrErr(msgStr); 00696 WriteMsg(msgStr,1); 00697 } 00698 00699 Dsize = readMap(filename,gTemp); 00700 00701 if(debug) { 00702 sprintf(msgStr,"name = %s, proc = %d, scale_value = %f, offset_value = %f, size = %x\n ", 00703 filename,procnum,scale_value,offset_value,Dsize); 00704 WriteMsg(msgStr,1); 00705 } 00706 for (i=1; i<=s0; i++) { 00707 for (j=1; j<=s1; j++) { 00708 k0 = Dsize*((i-1)*lcl_size[1] + (j-1)); 00709 tmpPtr = gTemp+k0; 00710 switch(Dsize) { 00711 case 1: temp = gTemp[k0]; break; 00712 case 2: temp = gTemp[k0]*256 + gTemp[k0+1]; break; 00713 case 3: temp = gTemp[k0]*65536 + gTemp[k0+1]*256 + gTemp[k0+2]; break; 00714 case 4: temp = gTemp[k0]*16777216 + gTemp[k0+1]*65536 + gTemp[k0+2]*256 + gTemp[k0+3]; break; 00715 default: fprintf(stderr,"ERROR, illegal size: %x\n",Dsize); temp = 0; 00716 } 00717 switch (Mtype) { 00718 case 'f' : 00719 ((float*)Map)[T(i,j)] = scale_value*((float)temp)+offset_value; 00720 break; 00721 case 'd' : case 'i' : 00722 ((int*)Map)[T(i,j)] = (int)(scale_value * (float)temp + offset_value); 00723 break; 00724 case 'c' : 00725 ((unsigned char*)Map)[T(i,j)] = (int)(scale_value * (unsigned char)temp); 00726 break; 00727 } 00728 } 00729 } 00730 if(debug) quick_look(Map, (char*)filename, Mtype, dbgPt.x, dbgPt.y, 2,'E'); 00731 link_edges(Map,Mtype); 00732 fflush(stderr); 00733 return Dsize; 00734 }
| int scan_forward | ( | FILE * | infile, | |
| const char * | tstring | |||
| ) |
Scan forward until a particular string is found.
- Parameters:
-
infile The file being read tstring The sought-after string
- Returns:
- Success/failure
Definition at line 1814 of file Driver_Utilities.c.
01815 { 01816 int sLen, i, cnt=0; 01817 char Input_string[100], test; 01818 01819 sLen = strlen(tstring); 01820 while( ( test = fgetc(infile) ) != EOF ) { 01821 for(i=0; i<(sLen-1); i++) 01822 Input_string[i] = Input_string[i+1]; 01823 Input_string[sLen-1] = test; 01824 Input_string[sLen] = '\0'; 01825 if(++cnt >= sLen) { 01826 test = strcmp(Input_string,tstring); 01827 if( test == 0 ) return 1; 01828 } 01829 } 01830 return(-1); 01831 }
| VOIDP nalloc | ( | unsigned | mem_size, | |
| const char | var_name[] | |||
| ) |
Allocate memory for a variable.
- Parameters:
-
mem_size The size of memory space var_name The variable's name
- Returns:
- Pointer to that memory
Definition at line 1857 of file Driver_Utilities.c.
01858 { 01859 VOIDP rp; 01860 01861 01862 if(mem_size == 0) return(NULL); 01863 rp = (VOIDP)malloc( mem_size ); 01864 total_memory += mem_size; 01865 fasync(stderr); 01866 if( rp == NULL ) { 01867 fprintf(stderr,"Sorry, out of memory(%d): %s\n",mem_size,var_name); 01868 Exit(0); 01869 } 01870 fmulti(stderr); 01871 return(rp); 01872 }
| float FMOD | ( | float | x, | |
| float | y | |||
| ) |
Modulus of a pair of (double) arguments.
- Parameters:
-
x Numerator y Denominator
- Returns:
- modulus (float) result
Definition at line 1675 of file Driver_Utilities.c.
| void PTSL_ReadLists | ( | PTSeriesList * | thisStruct, | |
| const char * | ptsFileName, | |||
| int | index, | |||
| float * | timeStep, | |||
| int * | nPtTS, | |||
| int | col | |||
| ) |
Point Time Series interpolation (unused): read raw point data.
Definition at line 1218 of file Driver_Utilities.c.
References broadcastInt(), debug, Exit(), gNPtTs, gTS, H_OPSYS, Lprocnum, modelFileName, ModelPath, msgStr, ProjName, PTSL_AddpTSeries(), UNIX, usrErr(), and WriteMsg().
01220 { 01221 char pathname[150], infilename[60], ss[201], ret = '\n'; 01222 FILE* cfile; 01223 int ix, iy, tst, sCnt=0; 01224 if( Lprocnum == 1 ) { 01225 if(H_OPSYS==UNIX) sprintf(modelFileName,"%s/%s/Data/%s.pts",ModelPath,ProjName,ptsFileName); 01226 else sprintf(modelFileName,"%s%s:Data:%s.pts",ModelPath,ProjName,ptsFileName); 01227 cfile = fopen(modelFileName,"r"); 01228 if(cfile==NULL) {fprintf(stdout,"\nERROR: Unable to open timeseries file %s\n",modelFileName); Exit(1); } 01229 else { sprintf(msgStr,"\nReading file %s\n",modelFileName); WriteMsg(msgStr,1); } 01230 if (debug > 2) {sprintf(msgStr,"Reading %s timeseries, column %d",modelFileName, col); usrErr(msgStr);} 01231 01232 01233 01234 fgets(ss,200,cfile); /* skip header line */ 01235 } 01236 while(1) { 01237 if( Lprocnum == 1 ) { 01238 tst = fscanf(cfile,"%d",&ix); 01239 if( tst > 0 ) { 01240 fscanf(cfile,"%d",&iy); 01241 tst = fscanf(cfile,"%s",infilename); 01242 sprintf(modelFileName,"%s/%s/Data/%s",ModelPath,ProjName,infilename); 01243 } 01244 } 01245 broadcastInt(&tst); 01246 if(tst<1) break; 01247 broadcastInt(&ix); 01248 broadcastInt(&iy); 01249 PTSL_AddpTSeries(thisStruct, ix, iy, index, sCnt, col); 01250 sCnt++; 01251 } 01252 if( Lprocnum == 1 ) fclose(cfile); 01253 *nPtTS = gNPtTs; 01254 *timeStep = gTS; 01255 }
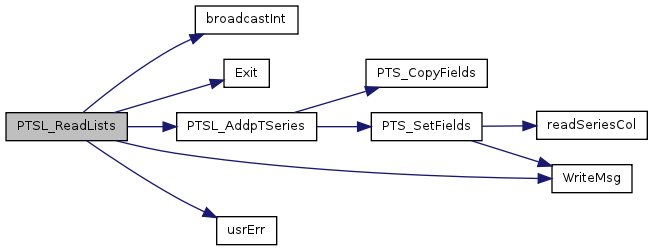
| void PTSL_CreatePointMap | ( | PTSeriesList * | pList, | |
| void * | Map, | |||
| unsigned char | Mtype, | |||
| int | step, | |||
| float | scale | |||
| ) |
Point Time Series interpolation (unused): generate interpolated spatial (map) data.
Definition at line 1258 of file Driver_Utilities.c.
References ON_MAP, PTSL_GetInterpolatedValue0(), s0, s1, and T.
01260 { 01261 int i0, i1; 01262 01263 switch(Mtype) { 01264 case 'f' : 01265 for(i0=0; i0<=s0+1; i0++) 01266 for(i1=0; i1<=s1+1; i1++) 01267 ((float*)Map)[T(i0,i1)] = (ON_MAP[T(i0,i1)]) ? PTSL_GetInterpolatedValue0(pList,i0,i1,step)*scale : 0.0 /*was this value (v0.5beta) -0.012 */; 01268 break; 01269 case 'i' : case 'd' : 01270 for(i0=0; i0<=s0+1; i0++) 01271 for(i1=0; i1<=s1+1; i1++) 01272 ((int*)Map)[T(i0,i1)] = (int) ( (ON_MAP[T(i0,i1)]) ? PTSL_GetInterpolatedValue0(pList,i0,i1,step)*scale : 0 ); 01273 break; 01274 case 'c' : 01275 for(i0=0; i0<=s0+1; i0++) 01276 for(i1=0; i1<=s1+1; i1++) 01277 ((unsigned char*)Map)[T(i0,i1)] = (unsigned char) '\0' + (int) ( (ON_MAP[T(i0,i1)]) ? PTSL_GetInterpolatedValue0(pList,i0,i1,step)*scale : 0 ); 01278 break; 01279 } 01280 }

| void stats | ( | int | step | ) |
Calling function for budget and summary stats.
Called every time step, this calls functions to summarize variables across time At the time interval for output, this calls functions to make the final summaries, and then resets the data for new interval.
Definition at line 63 of file BudgStats.c.
References BIRbudg_date(), BIRbudg_reset(), BIRbudg_sum(), BIRbudg_sumFinal(), BIRstats_date(), BIRstats_hydro_date(), BIRstats_hydro_reset(), BIRstats_hydro_sumFinal(), BIRstats_reset(), BIRstats_sum(), BIRstats_sumFinal(), CellAvg(), simTime::IsBIRavgEnd, simTime::IsBIRhydEnd, simTime::IsBudgEnd, simTime::IsBudgFirst, simTime::IsDay0, and SimTime.
00064 { 00065 CellAvg(); /* Generate sums, means (avgs), on cell-by-cell basis (not by Basin/IRegion here) */ 00066 00067 BIRstats_sum(); 00068 BIRbudg_sum(); 00069 00070 /* v2.8 hydro Perf Measure summaries - is a first cut, TODO is re-organize it */ 00071 if ( (SimTime.IsBIRhydEnd) ) { 00072 if (!SimTime.IsDay0) BIRstats_hydro_date(); 00073 BIRstats_hydro_sumFinal(); 00074 BIRstats_hydro_reset(); 00075 } 00076 if ( (SimTime.IsBIRavgEnd) ) { 00077 if (!SimTime.IsDay0) BIRstats_date(); /* skip TIME 0 of simulation */ 00078 BIRstats_sumFinal(); 00079 BIRstats_reset(); 00080 } 00081 if ( (SimTime.IsBudgEnd) ) { 00082 if (!SimTime.IsBudgFirst) BIRbudg_date(); /* skip first budget interval to accumulate info for mass-balance budget check */ 00083 BIRbudg_sumFinal(); 00084 BIRbudg_reset(); 00085 } 00086 }
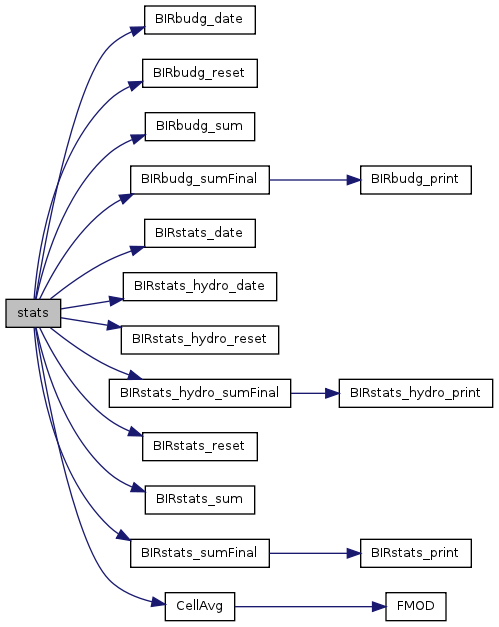
| void alloc_mem_stats | ( | void | ) |
Allocate memory for the BIR-based and cell-based stats variables.
Definition at line 2137 of file BudgStats.c.
References basn, BCmodel_sfwatAvg, BIR_DayFire, BIR_Sf_vel, C_Peri_mortAvg, C_Peri_nppAvg, C_Peri_PCAvg, C_PeriAvg, C_PeriNutCFAvg, C_PeriRespAvg, Calg_GPP, Calg_mort, CLsf_avg, Cperi_avg, DayFire, dop_decomp, dop_desorb, dop_macIn, dop_sorbIn, Elev_avg, ETAvg, EVAP, EvapAvg, floc_decomp, Floc_fr_phBioAvg, floc_In, HydPerAnn, HydRelDepPosNegAvg, init_pvar(), LAI_effAvg, Mac_avg, mac_mort, mac_nph_PCAvg, Mac_nphBioAvg, Mac_nphMortAvg, mac_NPP, Mac_nppAvg, mac_ph_PCAvg, Mac_phBioAvg, Mac_phMortAvg, Mac_totBioAvg, MacNutCfAvg, MacWatCfAvg, Manning_nAvg, msgStr, nalloc(), NC_Peri_mortAvg, NC_Peri_nppAvg, NC_Peri_PCAvg, NC_PeriAvg, NC_PeriNutCFAvg, NC_PeriRespAvg, NCalg_GPP, NCalg_mort, NCperi_avg, numBasn, numCells, ON_MAP, P, P_ATMOS, P_Calg, P_CELL, P_DEAD, P_DEAD_CELL, P_DEAD_ERR, P_DEAD_ERR_CUM, P_DEAD_IN, P_DEAD_IN_AVG, P_DEAD_IN_SUM, P_DEAD_OLD, P_DEAD_OUT, P_DEAD_OUT_AVG, P_DEAD_OUT_SUM, P_ERR, P_ERR_CUM, P_IN, P_IN_AVG, P_IN_GW, P_IN_OVL, P_IN_SPG, P_IN_STR, P_IN_SUM, P_LIVE, P_LIVE_CELL, P_LIVE_ERR, P_LIVE_ERR_CUM, P_LIVE_IN, P_LIVE_IN_AVG, P_LIVE_IN_SUM, P_LIVE_OLD, P_LIVE_OUT, P_LIVE_OUT_AVG, P_LIVE_OUT_SUM, P_MAC, P_NCalg, P_OLD, P_OUT, P_OUT_AVG, P_OUT_GW, P_OUT_OVL, P_OUT_SPG, P_OUT_STR, P_OUT_SUM, P_settl, P_WAT, P_WAT_CELL, P_WAT_ERR, P_WAT_ERR_CUM, P_WAT_IN, P_WAT_IN_AVG, P_WAT_IN_SUM, P_WAT_OLD, P_WAT_OUT, P_WAT_OUT_AVG, P_WAT_OUT_SUM, PeriAvg, PeriLiteCFAvg, RAIN, RainAvg, RCHG, s0, s1, S_ERR_CUM, S_IN, S_IN_AVG, S_IN_GW, S_IN_OVL, S_IN_SPG, S_IN_STR, S_IN_SUM, S_OUT, S_OUT_AVG, S_OUT_GW, S_OUT_OVL, S_OUT_SPG, S_OUT_STR, S_OUT_SUM, SALT_ATMOS, SaltSedAvg, SaltSfAvg, SedElevAvg, SF_WT_VEL_magAvg, Sfwat_avg, SfWatAvg, StgMinElev, SUMGW, SUMSF, SUMUW, T, TOT_P_CAN, TOT_S, TOT_S_CAN, TOT_S_CELL, TOT_S_ERR, TOT_S_OLD, TOT_VOL, TOT_VOL_AVG_ERR, TOT_VOL_CAN, TOT_VOL_CUM_ERR, TOT_VOL_ERR, TOT_VOL_OLD, TotHeadAvg, TP_settlAvg, TPpore_avg, TPSedMinAvg, TPSedUptAvg, TPSedWatAvg, TPsf_avg, TPSfMinAvg, TPSfUptAvg, TPSfWatAvg, TPsoil_avg, TPSorbAvg, TPtoSOILAvg, TPtoVOLAvg, TRANSP, TranspAvg, Unsat_avg, UnsatMoistAvg, UnsatZavg, usrErr0(), VOL_ERR_PER_INFL, VOL_IN, VOL_IN_AVG, VOL_IN_GW, VOL_IN_OVL, VOL_IN_SPG, VOL_IN_STR, VOL_IN_SUM, VOL_OUT, VOL_OUT_AVG, VOL_OUT_GW, VOL_OUT_OVL, VOL_OUT_SPG, VOL_OUT_STR, VOL_OUT_SUM, wat_sedMiner, wat_sedUpt, wat_sfMiner, and wat_sfUpt.
02138 { 02139 int ix,iy,cellLoc, numBasnMax=0; 02140 02141 02142 for(ix=0; ix<=s0+1; ix++) 02143 for(iy=0; iy<=s1+1; iy++) 02144 if (ON_MAP[cellLoc= T(ix,iy)]) { 02145 if (basn[cellLoc] > numBasnMax) numBasnMax = basn[cellLoc]; 02146 } 02147 numBasn = numBasnMax; /* total number of basins in domain */ 02148 02149 sprintf (msgStr," %d Basins/Indicator-Regions...",numBasn); usrErr0(msgStr); 02150 02151 /* basin budget arrays (number of actual basins plus basin 0, which is whole system) */ 02152 numCells = (int *) nalloc (sizeof(int)*(numBasn+1),"numCells"); /* number of cells in a basin */ 02153 02154 /* Indicator region averages (non-budget) */ 02155 /* v2.8 hydro Perf Measure summaries - is a first cut, TODO is re-organize it */ 02156 StgMinElev = (double *) nalloc(sizeof(double)*(numBasn+1),"StgMinElev"); 02157 init_pvar(StgMinElev,NULL,'b',0.0); 02158 BIR_Sf_vel = (double *) nalloc(sizeof(double)*(numBasn+1),"BIR_Sf_vel"); 02159 init_pvar(BIR_Sf_vel,NULL,'b',0.0); 02160 BIR_DayFire = (double *) nalloc(sizeof(double)*(numBasn+1),"BIR_DayFire"); 02161 init_pvar(BIR_DayFire,NULL,'b',0.0); 02162 DayFire = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"DayFire"); 02163 init_pvar(DayFire,NULL,'f',0.0); 02164 CLsf_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"CLsf_avg"); 02165 init_pvar(CLsf_avg,NULL,'b',0.0); 02166 /* end of v2.8 hydro Perf Measure summaries - is a first cut, TODO is re-organize it */ 02167 02168 Sfwat_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"Sfwat_avg"); 02169 init_pvar(Sfwat_avg,NULL,'b',0.0); 02170 Unsat_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"Unsat_avg"); 02171 init_pvar(Unsat_avg,NULL,'b',0.0); 02172 TPsf_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"TPsf_avg"); 02173 init_pvar(TPsf_avg,NULL,'b',0.0); 02174 TPpore_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"TPpore_avg"); 02175 init_pvar(TPpore_avg,NULL,'b',0.0); 02176 TPsoil_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"TPsoil_avg"); 02177 init_pvar(TPsoil_avg,NULL,'b',0.0); 02178 NCperi_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"NCperi_avg"); 02179 init_pvar(NCperi_avg,NULL,'b',0.0); 02180 Cperi_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"Cperi_avg"); 02181 init_pvar(Cperi_avg,NULL,'b',0.0); 02182 Mac_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"Mac_avg"); 02183 init_pvar(Mac_avg,NULL,'b',0.0); 02184 Elev_avg = (double *) nalloc(sizeof(double)*(numBasn+1),"Elev_avg"); 02185 init_pvar(Elev_avg,NULL,'b',0.0); 02186 /* hydro mass bal vars */ 02187 SUMSF = (double *) nalloc(sizeof(double)*(numBasn+1),"SUMSF"); 02188 init_pvar(SUMSF,NULL,'b',0.0); 02189 SUMGW = (double *) nalloc(sizeof(double)*(numBasn+1),"SUMGW"); 02190 init_pvar(SUMGW,NULL,'b',0.0); 02191 SUMUW = (double *) nalloc(sizeof(double)*(numBasn+1),"SUMUW"); 02192 init_pvar(SUMUW,NULL,'b',0.0); 02193 RAIN = (double *) nalloc(sizeof(double)*(numBasn+1),"RAIN"); 02194 init_pvar(RAIN,NULL,'b',0.0); 02195 EVAP = (double *) nalloc(sizeof(double)*(numBasn+1),"EVAP"); 02196 init_pvar(EVAP,NULL,'b',0.0); 02197 TRANSP = (double *) nalloc(sizeof(double)*(numBasn+1),"TRANSP"); 02198 init_pvar(TRANSP,NULL,'b',0.0); 02199 RCHG = (double *) nalloc(sizeof(double)*(numBasn+1),"RCHG"); 02200 init_pvar(RCHG,NULL,'b',0.0); 02201 02202 TOT_VOL = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_VOL"); 02203 init_pvar(TOT_VOL,NULL,'b',0.0); 02204 TOT_VOL_OLD = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_VOL_OLD"); 02205 init_pvar(TOT_VOL_OLD,NULL,'b',0.0); 02206 VOL_IN = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_IN"); 02207 init_pvar(VOL_IN,NULL,'b',0.0); 02208 VOL_OUT = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_OUT"); 02209 init_pvar(VOL_OUT,NULL,'b',0.0); 02210 TOT_VOL_ERR = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_VOL_ERR"); 02211 init_pvar(TOT_VOL_ERR,NULL,'b',0.0); 02212 TOT_VOL_CUM_ERR = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_VOL_CUM_ERR"); 02213 init_pvar(TOT_VOL_CUM_ERR,NULL,'b',0.0); 02214 TOT_VOL_AVG_ERR = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_VOL_AVG_ERR"); 02215 init_pvar(TOT_VOL_AVG_ERR,NULL,'b',0.0); 02216 VOL_ERR_PER_INFL = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_ERR_PER_INFL"); 02217 init_pvar(VOL_ERR_PER_INFL,NULL,'b',0.0); 02218 02219 VOL_IN_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_IN_SUM"); 02220 init_pvar(VOL_IN_SUM,NULL,'b',0.0); 02221 VOL_OUT_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_OUT_SUM"); 02222 init_pvar(VOL_OUT_SUM,NULL,'b',0.0); 02223 VOL_IN_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_IN_AVG"); 02224 init_pvar(VOL_IN_AVG,NULL,'b',0.0); 02225 VOL_OUT_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_OUT_AVG"); 02226 init_pvar(VOL_OUT_AVG,NULL,'b',0.0); 02227 02228 TOT_VOL_CAN = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_VOL_CAN"); 02229 init_pvar(TOT_VOL_CAN,NULL,'b',0.0); 02230 02231 VOL_IN_STR = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_IN_STR"); 02232 init_pvar(VOL_IN_STR,NULL,'b',0.0); 02233 VOL_IN_OVL = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_IN_OVL"); 02234 init_pvar(VOL_IN_OVL,NULL,'b',0.0); 02235 VOL_IN_SPG = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_IN_SPG"); 02236 init_pvar(VOL_IN_SPG,NULL,'b',0.0); 02237 VOL_IN_GW = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_IN_GW"); 02238 init_pvar(VOL_IN_GW,NULL,'b',0.0); 02239 02240 VOL_OUT_STR = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_OUT_STR"); 02241 init_pvar(VOL_OUT_STR,NULL,'b',0.0); 02242 VOL_OUT_OVL = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_OUT_OVL"); 02243 init_pvar(VOL_OUT_OVL,NULL,'b',0.0); 02244 VOL_OUT_SPG = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_OUT_SPG"); 02245 init_pvar(VOL_OUT_SPG,NULL,'b',0.0); 02246 VOL_OUT_GW = (double *) nalloc(sizeof(double)*(numBasn+1),"VOL_OUT_GW"); 02247 init_pvar(VOL_OUT_GW,NULL,'b',0.0); 02248 02249 /* phosph mass bal vars for all storage fractions */ 02250 P_CELL = (double *) nalloc(sizeof(double)*(numBasn+1),"P_CELL"); 02251 init_pvar(P_CELL,NULL,'b',0.0); 02252 P_ATMOS = (double *) nalloc(sizeof(double)*(numBasn+1),"P_ATMOS"); 02253 init_pvar(P_ATMOS,NULL,'b',0.0); 02254 02255 P = (double *) nalloc(sizeof(double)*(numBasn+1),"P"); 02256 init_pvar(P,NULL,'b',0.0); 02257 P_OLD = (double *) nalloc(sizeof(double)*(numBasn+1),"P_OLD"); 02258 init_pvar(P_OLD,NULL,'b',0.0); 02259 P_IN = (double *) nalloc(sizeof(double)*(numBasn+1),"P_IN"); 02260 init_pvar(P_IN,NULL,'b',0.0); 02261 P_OUT = (double *) nalloc(sizeof(double)*(numBasn+1),"P_OUT"); 02262 init_pvar(P_OUT,NULL,'b',0.0); 02263 P_ERR = (double *) nalloc(sizeof(double)*(numBasn+1),"P_ERR"); 02264 init_pvar(P_ERR,NULL,'b',0.0); 02265 P_ERR_CUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_ERR_CUM"); 02266 init_pvar(P_ERR_CUM,NULL,'b',0.0); 02267 02268 P_IN_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_IN_SUM"); 02269 init_pvar(P_IN_SUM,NULL,'b',0.0); 02270 P_OUT_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_OUT_SUM"); 02271 init_pvar(P_OUT_SUM,NULL,'b',0.0); 02272 P_IN_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_IN_AVG"); 02273 init_pvar(P_IN_AVG,NULL,'b',0.0); 02274 P_OUT_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_OUT_AVG"); 02275 init_pvar(P_OUT_AVG,NULL,'b',0.0); 02276 02277 TOT_P_CAN = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_P_CAN"); 02278 init_pvar(TOT_P_CAN,NULL,'b',0.0); 02279 02280 P_IN_STR = (double *) nalloc(sizeof(double)*(numBasn+1),"P_IN_STR"); 02281 init_pvar(P_IN_STR,NULL,'b',0.0); 02282 P_IN_OVL = (double *) nalloc(sizeof(double)*(numBasn+1),"P_IN_OVL"); 02283 init_pvar(P_IN_OVL,NULL,'b',0.0); 02284 P_IN_SPG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_IN_SPG"); 02285 init_pvar(P_IN_SPG,NULL,'b',0.0); 02286 P_IN_GW = (double *) nalloc(sizeof(double)*(numBasn+1),"P_IN_GW"); 02287 init_pvar(P_IN_GW,NULL,'b',0.0); 02288 02289 P_OUT_STR = (double *) nalloc(sizeof(double)*(numBasn+1),"P_OUT_STR"); 02290 init_pvar(P_OUT_STR,NULL,'b',0.0); 02291 P_OUT_OVL = (double *) nalloc(sizeof(double)*(numBasn+1),"P_OUT_OVL"); 02292 init_pvar(P_OUT_OVL,NULL,'b',0.0); 02293 P_OUT_SPG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_OUT_SPG"); 02294 init_pvar(P_OUT_SPG,NULL,'b',0.0); 02295 P_OUT_GW = (double *) nalloc(sizeof(double)*(numBasn+1),"P_OUT_GW"); 02296 init_pvar(P_OUT_GW,NULL,'b',0.0); 02297 02298 /* phosph mass bal vars for live fraction */ 02299 P_LIVE_CELL = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_CELL"); 02300 init_pvar(P_LIVE_CELL,NULL,'b',0.0); 02301 P_MAC = (double *) nalloc(sizeof(double)*(numBasn+1),"P_MAC"); 02302 init_pvar(P_MAC,NULL,'b',0.0); 02303 P_Calg = (double *) nalloc(sizeof(double)*(numBasn+1),"P_Calg"); 02304 init_pvar(P_Calg,NULL,'b',0.0); 02305 P_NCalg = (double *) nalloc(sizeof(double)*(numBasn+1),"P_NCalg"); 02306 init_pvar(P_NCalg,NULL,'b',0.0); 02307 Calg_GPP = (double *) nalloc(sizeof(double)*(numBasn+1),"Calg_GPP"); 02308 init_pvar(Calg_GPP,NULL,'b',0.0); 02309 NCalg_GPP = (double *) nalloc(sizeof(double)*(numBasn+1),"NCalg_GPP"); 02310 init_pvar(NCalg_GPP,NULL,'b',0.0); 02311 Calg_mort = (double *) nalloc(sizeof(double)*(numBasn+1),"Calg_mort"); 02312 init_pvar(Calg_mort,NULL,'b',0.0); 02313 NCalg_mort = (double *) nalloc(sizeof(double)*(numBasn+1),"NCalg_mort"); 02314 init_pvar(NCalg_mort,NULL,'b',0.0); 02315 02316 mac_NPP = (double *) nalloc(sizeof(double)*(numBasn+1),"mac_NPP"); 02317 init_pvar(mac_NPP,NULL,'b',0.0); 02318 mac_mort = (double *) nalloc(sizeof(double)*(numBasn+1),"mac_mort"); 02319 init_pvar(mac_mort,NULL,'b',0.0); 02320 02321 P_LIVE = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE"); 02322 init_pvar(P_LIVE,NULL,'b',0.0); 02323 02324 P_LIVE_OLD = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_OLD"); 02325 init_pvar(P_LIVE_OLD,NULL,'b',0.0); 02326 P_LIVE_IN = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_IN"); 02327 init_pvar(P_LIVE_IN,NULL,'b',0.0); 02328 P_LIVE_OUT = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_OUT"); 02329 init_pvar(P_LIVE_OUT,NULL,'b',0.0); 02330 P_LIVE_ERR = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_ERR"); 02331 init_pvar(P_LIVE_ERR,NULL,'b',0.0); 02332 P_LIVE_ERR_CUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_ERR_CUM"); 02333 init_pvar(P_LIVE_ERR_CUM,NULL,'b',0.0); 02334 02335 P_LIVE_IN_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_IN_SUM"); 02336 init_pvar(P_LIVE_IN_SUM,NULL,'b',0.0); 02337 P_LIVE_OUT_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_OUT_SUM"); 02338 init_pvar(P_LIVE_OUT_SUM,NULL,'b',0.0); 02339 P_LIVE_IN_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_IN_AVG"); 02340 init_pvar(P_LIVE_IN_AVG,NULL,'b',0.0); 02341 P_LIVE_OUT_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_LIVE_OUT_AVG"); 02342 init_pvar(P_LIVE_OUT_AVG,NULL,'b',0.0); 02343 02344 /* phosph mass bal vars for dead fraction */ 02345 P_DEAD_CELL = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_CELL"); 02346 init_pvar(P_DEAD_CELL,NULL,'b',0.0); 02347 dop_macIn = (double *) nalloc(sizeof(double)*(numBasn+1),"dop_macIn"); 02348 init_pvar(dop_macIn,NULL,'b',0.0); 02349 dop_sorbIn = (double *) nalloc(sizeof(double)*(numBasn+1),"dop_sorbIn"); 02350 init_pvar(dop_sorbIn,NULL,'b',0.0); 02351 dop_decomp = (double *) nalloc(sizeof(double)*(numBasn+1),"dop_decomp"); 02352 init_pvar(dop_decomp,NULL,'b',0.0); 02353 dop_desorb = (double *) nalloc(sizeof(double)*(numBasn+1),"dop_desorb"); 02354 init_pvar(dop_desorb,NULL,'b',0.0); 02355 floc_decomp = (double *) nalloc(sizeof(double)*(numBasn+1),"floc_decomp"); 02356 init_pvar(floc_decomp,NULL,'b',0.0); 02357 floc_In = (double *) nalloc(sizeof(double)*(numBasn+1),"floc_In"); 02358 init_pvar(floc_In,NULL,'b',0.0); 02359 02360 P_DEAD = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD"); 02361 init_pvar(P_DEAD,NULL,'b',0.0); 02362 02363 P_DEAD_OLD = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_OLD"); 02364 init_pvar(P_DEAD_OLD,NULL,'b',0.0); 02365 P_DEAD_IN = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_IN"); 02366 init_pvar(P_DEAD_IN,NULL,'b',0.0); 02367 P_DEAD_OUT = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_OUT"); 02368 init_pvar(P_DEAD_OUT,NULL,'b',0.0); 02369 P_DEAD_ERR = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_ERR"); 02370 init_pvar(P_DEAD_ERR,NULL,'b',0.0); 02371 P_DEAD_ERR_CUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_ERR_CUM"); 02372 init_pvar(P_DEAD_ERR_CUM,NULL,'b',0.0); 02373 02374 P_DEAD_IN_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_IN_SUM"); 02375 init_pvar(P_DEAD_IN_SUM,NULL,'b',0.0); 02376 P_DEAD_OUT_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_OUT_SUM"); 02377 init_pvar(P_DEAD_OUT_SUM,NULL,'b',0.0); 02378 P_DEAD_IN_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_IN_AVG"); 02379 init_pvar(P_DEAD_IN_AVG,NULL,'b',0.0); 02380 P_DEAD_OUT_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_DEAD_OUT_AVG"); 02381 init_pvar(P_DEAD_OUT_AVG,NULL,'b',0.0); 02382 02383 /* phosph mass bal vars for water-borne fraction */ 02384 P_WAT_CELL = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_CELL"); 02385 init_pvar(P_WAT_CELL,NULL,'b',0.0); 02386 wat_sfMiner = (double *) nalloc(sizeof(double)*(numBasn+1),"wat_sfMiner"); 02387 init_pvar(wat_sfMiner,NULL,'b',0.0); 02388 wat_sedMiner = (double *) nalloc(sizeof(double)*(numBasn+1),"wat_sedMiner"); 02389 init_pvar(wat_sedMiner,NULL,'b',0.0); 02390 wat_sfUpt = (double *) nalloc(sizeof(double)*(numBasn+1),"wat_sfUpt"); 02391 init_pvar(wat_sfUpt,NULL,'b',0.0); 02392 P_settl = (double *) nalloc(sizeof(double)*(numBasn+1),"P_settl"); 02393 init_pvar(P_settl,NULL,'b',0.0); 02394 wat_sedUpt = (double *) nalloc(sizeof(double)*(numBasn+1),"wat_sedUpt"); 02395 init_pvar(wat_sedUpt,NULL,'b',0.0); 02396 02397 P_WAT = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT"); 02398 init_pvar(P_WAT,NULL,'b',0.0); 02399 02400 P_WAT_OLD = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_OLD"); 02401 init_pvar(P_WAT_OLD,NULL,'b',0.0); 02402 P_WAT_IN = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_IN"); 02403 init_pvar(P_WAT_IN,NULL,'b',0.0); 02404 P_WAT_OUT = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_OUT"); 02405 init_pvar(P_WAT_OUT,NULL,'b',0.0); 02406 P_WAT_ERR = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_ERR"); 02407 init_pvar(P_WAT_ERR,NULL,'b',0.0); 02408 P_WAT_ERR_CUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_ERR_CUM"); 02409 init_pvar(P_WAT_ERR_CUM,NULL,'b',0.0); 02410 02411 P_WAT_IN_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_IN_SUM"); 02412 init_pvar(P_WAT_IN_SUM,NULL,'b',0.0); 02413 P_WAT_OUT_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_OUT_SUM"); 02414 init_pvar(P_WAT_OUT_SUM,NULL,'b',0.0); 02415 P_WAT_IN_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_IN_AVG"); 02416 init_pvar(P_WAT_IN_AVG,NULL,'b',0.0); 02417 P_WAT_OUT_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"P_WAT_OUT_AVG"); 02418 init_pvar(P_WAT_OUT_AVG,NULL,'b',0.0); 02419 02420 /* salt mass bal vars */ 02421 TOT_S_CELL = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_S_CELL"); 02422 init_pvar(TOT_S_CELL,NULL,'b',0.0); 02423 SALT_ATMOS = (double *) nalloc(sizeof(double)*(numBasn+1),"SALT_ATMOS"); 02424 init_pvar(SALT_ATMOS,NULL,'b',0.0); 02425 02426 TOT_S = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_S"); 02427 init_pvar(TOT_S,NULL,'b',0.0); 02428 TOT_S_OLD = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_S_OLD"); 02429 init_pvar(TOT_S_OLD,NULL,'b',0.0); 02430 S_IN = (double *) nalloc(sizeof(double)*(numBasn+1),"S_IN"); 02431 init_pvar(S_IN,NULL,'b',0.0); 02432 S_OUT = (double *) nalloc(sizeof(double)*(numBasn+1),"S_OUT"); 02433 init_pvar(S_OUT,NULL,'b',0.0); 02434 TOT_S_ERR = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_S_ERR"); 02435 init_pvar(TOT_S_ERR,NULL,'b',0.0); 02436 S_ERR_CUM = (double *) nalloc(sizeof(double)*(numBasn+1),"S_ERR_CUM"); 02437 init_pvar(S_ERR_CUM,NULL,'b',0.0); 02438 02439 S_IN_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"S_IN_SUM"); 02440 init_pvar(S_IN_SUM,NULL,'b',0.0); 02441 S_OUT_SUM = (double *) nalloc(sizeof(double)*(numBasn+1),"S_OUT_SUM"); 02442 init_pvar(S_OUT_SUM,NULL,'b',0.0); 02443 S_IN_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"S_IN_AVG"); 02444 init_pvar(S_IN_AVG,NULL,'b',0.0); 02445 S_OUT_AVG = (double *) nalloc(sizeof(double)*(numBasn+1),"S_OUT_AVG"); 02446 init_pvar(S_OUT_AVG,NULL,'b',0.0); 02447 02448 TOT_S_CAN = (double *) nalloc(sizeof(double)*(numBasn+1),"TOT_S_CAN"); 02449 init_pvar(TOT_S_CAN,NULL,'b',0.0); 02450 02451 S_IN_STR = (double *) nalloc(sizeof(double)*(numBasn+1),"S_IN_STR"); 02452 init_pvar(S_IN_STR,NULL,'b',0.0); 02453 S_IN_OVL = (double *) nalloc(sizeof(double)*(numBasn+1),"S_IN_OVL"); 02454 init_pvar(S_IN_OVL,NULL,'b',0.0); 02455 S_IN_SPG = (double *) nalloc(sizeof(double)*(numBasn+1),"S_IN_SPG"); 02456 init_pvar(S_IN_SPG,NULL,'b',0.0); 02457 S_IN_GW = (double *) nalloc(sizeof(double)*(numBasn+1),"S_IN_GW"); 02458 init_pvar(S_IN_GW,NULL,'b',0.0); 02459 02460 S_OUT_STR = (double *) nalloc(sizeof(double)*(numBasn+1),"S_OUT_STR"); 02461 init_pvar(S_OUT_STR,NULL,'b',0.0); 02462 S_OUT_OVL = (double *) nalloc(sizeof(double)*(numBasn+1),"S_OUT_OVL"); 02463 init_pvar(S_OUT_OVL,NULL,'b',0.0); 02464 S_OUT_SPG = (double *) nalloc(sizeof(double)*(numBasn+1),"S_OUT_SPG"); 02465 init_pvar(S_OUT_SPG,NULL,'b',0.0); 02466 S_OUT_GW = (double *) nalloc(sizeof(double)*(numBasn+1),"S_OUT_GW"); 02467 init_pvar(S_OUT_GW,NULL,'b',0.0); 02468 02469 /* map arrays with summary stats */ 02470 HydPerAnn = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HydPerAnn"); 02471 init_pvar(HydPerAnn,NULL,'f',0.0); 02472 SfWatAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SfWatAvg"); 02473 init_pvar(SfWatAvg,NULL,'f',0.0); 02474 TotHeadAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TotHeadAvg"); 02475 init_pvar(TotHeadAvg,NULL,'f',0.0); 02476 HydRelDepPosNegAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"HydRelDepPosNegAvg"); 02477 init_pvar(HydRelDepPosNegAvg,NULL,'f',0.0); 02478 UnsatZavg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UnsatZavg"); 02479 init_pvar(UnsatZavg,NULL,'f',0.0); 02480 02481 RainAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"RainAvg"); 02482 init_pvar(RainAvg,NULL,'f',0.0); 02483 ETAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"ETAvg"); 02484 init_pvar(ETAvg,NULL,'f',0.0); 02485 EvapAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"EvapAvg"); 02486 init_pvar(EvapAvg,NULL,'f',0.0); 02487 TranspAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TranspAvg"); 02488 init_pvar(TranspAvg,NULL,'f',0.0); 02489 UnsatMoistAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"UnsatMoistAvg"); 02490 init_pvar(UnsatMoistAvg,NULL,'f',0.0); 02491 /* v2.7.0 addition */ 02492 SF_WT_VEL_magAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SF_WT_VEL_magAvg"); 02493 init_pvar(SF_WT_VEL_magAvg,NULL,'f',0.0); 02494 /* v2.7.1 addition */ 02495 BCmodel_sfwatAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"BCmodel_sfwatAvg"); 02496 init_pvar(BCmodel_sfwatAvg,NULL,'f',0.0); 02497 02498 TPSfWatAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPSfWatAvg"); 02499 init_pvar(TPSfWatAvg,NULL,'f',0.0); 02500 TPSfUptAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPSfUptAvg"); 02501 init_pvar(TPSfUptAvg,NULL,'f',0.0); 02502 TPSfMinAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPSfMinAvg"); 02503 init_pvar(TPSfMinAvg,NULL,'f',0.0); 02504 TP_settlAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TP_settlAvg"); 02505 init_pvar(TP_settlAvg,NULL,'f',0.0); 02506 TPSedWatAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPSedWatAvg"); 02507 init_pvar(TPSedWatAvg,NULL,'f',0.0); 02508 TPSedUptAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPSedUptAvg"); 02509 init_pvar(TPSedUptAvg,NULL,'f',0.0); 02510 TPSedMinAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPSedMinAvg"); 02511 init_pvar(TPSedMinAvg,NULL,'f',0.0); 02512 TPSorbAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPSorbAvg"); 02513 init_pvar(TPSorbAvg,NULL,'f',0.0); 02514 TPtoVOLAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoVOLAvg"); 02515 init_pvar(TPtoVOLAvg,NULL,'f',0.0); 02516 TPtoSOILAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"TPtoSOILAvg"); 02517 init_pvar(TPtoSOILAvg,NULL,'f',0.0); 02518 02519 SaltSfAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SaltSfAvg"); 02520 init_pvar(SaltSfAvg,NULL,'f',0.0); 02521 SaltSedAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SaltSedAvg"); 02522 init_pvar(SaltSedAvg,NULL,'f',0.0); 02523 02524 Floc_fr_phBioAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Floc_fr_phBioAvg"); 02525 init_pvar(Floc_fr_phBioAvg,NULL,'f',0.0); 02526 02527 NC_PeriAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_PeriAvg"); 02528 init_pvar(NC_PeriAvg,NULL,'f',0.0); 02529 NC_Peri_nppAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_Peri_nppAvg"); 02530 init_pvar(NC_Peri_nppAvg,NULL,'f',0.0); 02531 NC_Peri_mortAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_Peri_mortAvg"); 02532 init_pvar(NC_Peri_mortAvg,NULL,'f',0.0); 02533 NC_PeriRespAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_PeriRespAvg"); 02534 init_pvar(NC_PeriRespAvg,NULL,'f',0.0); 02535 NC_PeriNutCFAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_PeriNutCFAvg"); 02536 init_pvar(NC_PeriNutCFAvg,NULL,'f',0.0); 02537 NC_Peri_PCAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"NC_Peri_PCAvg"); 02538 init_pvar(NC_Peri_PCAvg,NULL,'f',0.0); 02539 C_PeriAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_PeriAvg"); 02540 init_pvar(C_PeriAvg,NULL,'f',0.0); 02541 C_Peri_nppAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_Peri_nppAvg"); 02542 init_pvar(C_Peri_nppAvg,NULL,'f',0.0); 02543 C_Peri_mortAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_Peri_mortAvg"); 02544 init_pvar(C_Peri_mortAvg,NULL,'f',0.0); 02545 C_PeriRespAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_PeriRespAvg"); 02546 init_pvar(C_PeriRespAvg,NULL,'f',0.0); 02547 C_PeriNutCFAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_PeriNutCFAvg"); 02548 init_pvar(C_PeriNutCFAvg,NULL,'f',0.0); 02549 C_Peri_PCAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"C_Peri_PCAvg"); 02550 init_pvar(C_Peri_PCAvg,NULL,'f',0.0); 02551 PeriAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PeriAvg"); 02552 init_pvar(PeriAvg,NULL,'f',0.0); 02553 PeriLiteCFAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"PeriLiteCFAvg"); 02554 init_pvar(PeriLiteCFAvg,NULL,'f',0.0); 02555 02556 Mac_nphBioAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Mac_nphBioAvg"); 02557 init_pvar(Mac_nphBioAvg,NULL,'f',0.0); 02558 Mac_phBioAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Mac_phBioAvg"); 02559 init_pvar(Mac_phBioAvg,NULL,'f',0.0); 02560 Mac_totBioAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Mac_totBioAvg"); 02561 init_pvar(Mac_totBioAvg,NULL,'f',0.0); 02562 Mac_nppAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Mac_nppAvg"); 02563 init_pvar(Mac_nppAvg,NULL,'f',0.0); 02564 Mac_nphMortAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Mac_nphMortAvg"); 02565 init_pvar(Mac_nphMortAvg,NULL,'f',0.0); 02566 Mac_phMortAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Mac_phMortAvg"); 02567 init_pvar(Mac_phMortAvg,NULL,'f',0.0); 02568 LAI_effAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"LAI_effAvg"); 02569 init_pvar(LAI_effAvg,NULL,'f',0.0); 02570 Manning_nAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"Manning_nAvg"); 02571 init_pvar(Manning_nAvg,NULL,'f',0.0); 02572 MacNutCfAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MacNutCfAvg"); 02573 init_pvar(MacNutCfAvg,NULL,'f',0.0); 02574 MacWatCfAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"MacWatCfAvg"); 02575 init_pvar(MacWatCfAvg,NULL,'f',0.0); 02576 mac_nph_PCAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"mac_nph_PCAvg"); 02577 init_pvar(mac_nph_PCAvg,NULL,'f',0.0); 02578 mac_ph_PCAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"mac_ph_PCAvg"); 02579 init_pvar(mac_ph_PCAvg,NULL,'f',0.0); 02580 02581 SedElevAvg = (float *) nalloc(sizeof(float)*(s0+2)*(s1+2),"SedElevAvg"); 02582 init_pvar(SedElevAvg,NULL,'f',0.0); 02583 02584 } /* end of alloc_mem_stats */
| void BIRinit | ( | void | ) |
Set up the Basin & Indicator Region (BIR) linkages/inheritances.
Definition at line 1164 of file BudgStats.c.
References basInFile, basins, basn, basn_list, basndef::basnTxt, CELL_SIZE, conv_kgTOmg, conv_mTOcm, basndef::family, basndef::FLok, basndef::IR, modelFileName, modelName, ModelPath, msgStr, numBasn, numCells, basndef::numFLok, basndef::numIR, ON_MAP, basndef::parent, ProjName, s0, s1, Scip(), T, and usrErr().
01165 { 01166 int ix,iy,cellLoc,ibas; 01167 int ii, jj, basnID; 01168 int basnCnt=-1; /* the whole system basin-0 in the basinIR file is not considered part of this count of the number of Basin/IRs */ 01169 char ss[222], *line, modnam[20], boundIR[3]; 01170 01171 sprintf( modelFileName, "%s/%s/Data/basinIR", ModelPath, ProjName ); 01172 /* Open file with basin linkage/inheritance data */ 01173 if ( ( basInFile = fopen( modelFileName, "r" ) ) == NULL ) 01174 { 01175 sprintf( msgStr,"Can't open %s basin definition input file! ",modelFileName ) ; usrErr(msgStr); 01176 exit(-1) ; 01177 } 01178 01179 fgets( ss, 220, basInFile );fgets( ss, 220, basInFile ); /* skip 2 header lines */ 01180 fgets( ss, 220, basInFile ); sscanf( ss,"%s", &modnam); 01181 if (strcmp(modnam,modelName) != 0) { 01182 sprintf(msgStr, "The model name (%s) found in the %s file doesn't match the one (%s) you asked for in Driver.parm!", 01183 modnam, modelFileName, modelName); usrErr(msgStr); 01184 exit(-1); 01185 } 01186 /* Allocate memory for first basin */ 01187 if ( (basins = ( basnDef *) malloc( (size_t) sizeof( basnDef ))) == NULL ) { 01188 printf( "Failed to allocate memory for first basin (%s)\n ", basins->basnTxt ) ; 01189 exit( -2 ) ; 01190 } 01191 /* allocate memory for array of pointers to basin atts */ 01192 if ( (basn_list = 01193 ( basnDef **) malloc( (size_t) sizeof( basnDef *) * (numBasn+1))) == NULL ) 01194 { 01195 printf( "Failed to allocate memory for basin_list\n " ) ; 01196 exit( -2 ) ; 01197 }; 01198 01199 01200 fgets( ss, 220, basInFile ); /* skip the column name header */ 01201 01202 while ( fgets( ss, 220, basInFile ) != NULL && !feof( basInFile ) ) 01203 { 01204 line = ss; 01205 sscanf (line, "%d\t%s\t%d",&basnID, &basins->basnTxt,&basins->numIR); 01206 line = Scip( line, '\t' );line = Scip( line, '\t' );line = Scip( line, '\t' ); 01207 /* the indicator regions within a hydrologic basin are subscripted starting at 1, 01208 with the 0'th indicator region being the hydrologic basin itself. Flok is 01209 used to check for allowable flows among basins/indicator-regions, to determine if user configured 01210 BIRs are OK (NOT used to constrain calculated flows, but only used to verify that no unallowed interbasin 01211 overland flow is occuring via some "leak" in the vector topology) */ 01212 for (ii=0; ii<basins->numIR; ii++) { 01213 sscanf (line, "%d%", &basins->IR[ii]); 01214 basins->FLok[ii] = basins->IR[ii]; /* flow is allowed among IRegions in a particular hydro basin (family) */ 01215 basins->numFLok++; 01216 line = Scip( line, ',' ); 01217 } 01218 basnCnt++; /* count the number of basins&IRs to check that the # in the file is same as # defined on the map */ 01219 01220 /* if there are any FlowOk Basin/Indicator-Region attached to this Basin/Indicator-Region, read them */ 01221 while (1) { 01222 while ( *line != '#' && *line != '\0' ) line++; 01223 if(*line != '\0') { 01224 ++line; 01225 sscanf (line, "%d%", &basins->FLok[ii]); 01226 basins->numFLok++; 01227 ii++; 01228 } 01229 else break; 01230 } 01231 01232 basn_list[basnID] = basins; 01233 01234 /* Allocate memory for next basin */ 01235 if ( ( basins = ( basnDef *) malloc( (size_t) sizeof( basnDef ))) == NULL ) 01236 { 01237 printf( "Failed to allocate memory for next basin (%s)\n ", basins->basnTxt ) ; 01238 exit( -2 ) ; 01239 } 01240 01241 01242 } /* end of reading basin lines */ 01243 01244 free ((char *)basins); 01245 fclose(basInFile); 01246 01247 if (basnCnt != numBasn) { sprintf(msgStr, "Error - the %d basins read from basinIR does not match the %d basins in the basins map! Please fix the basinIR file.\n", 01248 basnCnt,numBasn); usrErr(msgStr); exit (-1); } 01249 01250 /* count the number of cells in the Basins/Indicator-Regions */ 01251 for (ibas=numBasn;ibas>=0;ibas--) numCells[ibas] = 0; 01252 for(ix=0; ix<=s0+1; ix++) 01253 for(iy=0; iy<=s1+1; iy++) 01254 if (ON_MAP[cellLoc= T(ix,iy)]) { 01255 numCells[basn[cellLoc]]++; /* count the number of cells in each basin */ 01256 numCells[0]++; /* count the number of cells in each basin */ 01257 } 01258 /* whole system (basin 0) is calc'd independently, while each of the hydro basins are 01259 sums of their respective Indicator Regions */ 01260 for (ibas=numBasn;ibas>=0;ibas--) { 01261 basins = basn_list[ibas]; 01262 basins->family = ibas; /* set the basin to initially be it's own family; below it may find that it is a child of a diff family */ 01263 basins->parent = ibas; /* set the basin to initially be a childless parent */ 01264 for (ii=0; ii<basins->numIR; ii++) { 01265 01266 basn_list[basins->IR[ii]]->family = ibas; /* the hydrologic basin family name of this IRegion */ 01267 basn_list[basins->IR[ii]]->parent = 0; /* this IR is a child, not a parent */ 01268 numCells[ibas] += numCells[basins->IR[ii]]; /* add the IRegion cells to the parent basin #cells */ 01269 } 01270 basins->conv_m3TOcm = conv_mTOcm/(numCells[ibas]*CELL_SIZE); 01271 basins->conv_kgTOmgm2 = conv_kgTOmg/(numCells[ibas]*CELL_SIZE); 01272 01273 } 01274 } /* end of BIRinit() */
| void BIRoutfiles | ( | void | ) |
Open files and create headers for BIR output.
Definition at line 1282 of file BudgStats.c.
References BIRavg1, BIRavg2, BIRavg3, BIRavg4, BIRavg5, BIRhydro, budget_P1, budget_P2, budget_P3, budget_P4, budget_P5, budget_Par1, budget_Par2, budget_Par3, budget_Par4, budget_Par5, budget_Pdead1, budget_Pdead2, budget_Pdead3, budget_Pdead4, budget_Pdead5, budget_Plive1, budget_Plive2, budget_Plive3, budget_Plive4, budget_Plive5, budget_Pwat1, budget_Pwat2, budget_Pwat3, budget_Pwat4, budget_Pwat5, budget_S1, budget_S2, budget_S3, budget_S4, budget_S5, budget_Wacr1, budget_Wacr2, budget_Wacr3, budget_Wacr4, budget_Wacr5, budget_Wcm1, budget_Wcm2, budget_Wcm3, budget_Wcm4, budget_Wcm5, CELL_SIZE, ESPmodeON, Min, modelFileName, modelName, modelVers, numBasn, numCells, OutputPath, ProjName, SimAlt, and SimModif.
01283 { 01284 int Fnum,Fsets,ibasLow,ibasHigh,ibas; 01285 01286 01287 01288 /********/ 01289 /* For the budget & BIRavg files, we can have multiple sets of files, with 01290 >1 sets needed if a large (>12) number of Basins/Indicator-Regions are defined. First, 01291 we open the files, then go to write headers for them */ 01292 01293 {/*Open the primary files for all of the budgets & BIRavgs */ 01294 01295 /* v2.8 hydro Perf Measure summaries - is a first cut, TODO is re-organize it */ 01296 sprintf( modelFileName, "%s/%s/Output/Budget/BIRhydro", OutputPath, ProjName ); 01297 if ( ( BIRhydro = fopen( modelFileName, "w" ) ) == NULL ) 01298 {printf( "Can't open BIRhydro file! " );exit(-1) ;} 01299 01300 /* Indicator region avgs */ 01301 sprintf( modelFileName, "%s/%s/Output/Budget/BIRavg1", OutputPath, ProjName ); 01302 if ( ( BIRavg1 = fopen( modelFileName, "w" ) ) == NULL ) 01303 {printf( "Can't open BIRavg1 file! " );exit(-1) ;} 01304 /* water budget/massCheck info - units in acre-feet */ 01305 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wacr1", OutputPath, ProjName ); 01306 if ( ( budget_Wacr1 = fopen( modelFileName, "w" ) ) == NULL ) 01307 {printf( "Can't open budg_Wacr1 file! " );exit(-1) ;} 01308 /* water budget/massCheck info - units in cm height */ 01309 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wcm1", OutputPath, ProjName ); 01310 if ( ( budget_Wcm1 = fopen( modelFileName, "w" ) ) == NULL ) 01311 {printf( "Can't open budg_Wcm1 file! " );exit(-1) ;} 01312 /* Phosph budget/massCheck info for all P fractions (units=Mg) */ 01313 sprintf( modelFileName, "%s/%s/Output/Budget/budg_P1", OutputPath, ProjName ); 01314 if ( ( budget_P1 = fopen( modelFileName, "w" ) ) == NULL ) 01315 {printf( "Can't open budg_P1 file! " );exit(-1) ;} 01316 /* Phosph budget/massCheck info for all P fractions (units=mg/m2) */ 01317 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Par1", OutputPath, ProjName ); 01318 if ( ( budget_Par1 = fopen( modelFileName, "w" ) ) == NULL ) 01319 {printf( "Can't open budg_Par1 file! " );exit(-1) ;} 01320 /* Salt budget/massCheck info */ 01321 sprintf( modelFileName, "%s/%s/Output/Budget/budg_S1", OutputPath, ProjName ); 01322 if ( ( budget_S1 = fopen( modelFileName, "w" ) ) == NULL ) 01323 {printf( "Can't open budg_S1 file! " );exit(-1) ;} 01324 if (!ESPmodeON) { 01325 /* Phosph budget/massCheck info for live P fraction */ 01326 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pliv1", OutputPath, ProjName ); 01327 if ( ( budget_Plive1 = fopen( modelFileName, "w" ) ) == NULL ) 01328 {printf( "Can't open budg_Pliv1 file! " );exit(-1) ;} 01329 /* Phosph budget/massCheck info for dead P fraction */ 01330 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pded1", OutputPath, ProjName ); 01331 if ( ( budget_Pdead1 = fopen( modelFileName, "w" ) ) == NULL ) 01332 {printf( "Can't open budg_Pded1 file! " );exit(-1) ;} 01333 /* Phosph budget/massCheck info for water-borne P fraction */ 01334 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pwat1", OutputPath, ProjName ); 01335 if ( ( budget_Pwat1 = fopen( modelFileName, "w" ) ) == NULL ) 01336 {printf( "Can't open budg_Pwat1 file! " );exit(-1) ;} 01337 } 01338 01339 01340 if (numBasn>=13) { 01341 /*Open the secondary files for all of the budgets & BIRavgs if we have more than 12 basins */ 01342 /* Indicator region avgs */ 01343 sprintf( modelFileName, "%s/%s/Output/Budget/BIRavg2", OutputPath, ProjName ); 01344 if ( ( BIRavg2 = fopen( modelFileName, "w" ) ) == NULL ) 01345 { printf( "Can't open BIRavg2 file! " ); exit(-1) ;} 01346 /* water budget/massCheck info - units in acre-feet */ 01347 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wacr2", OutputPath, ProjName ); 01348 if ( ( budget_Wacr2 = fopen( modelFileName, "w" ) ) == NULL ) 01349 { printf( "Can't open budg_Wacr2 file! " ); exit(-1) ;} 01350 /* water budget/massCheck info - units in cm height */ 01351 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wcm2", OutputPath, ProjName ); 01352 if ( ( budget_Wcm2 = fopen( modelFileName, "w" ) ) == NULL ) 01353 {printf( "Can't open budg_Wcm2 file! " ); exit(-1) ;} 01354 /* Phosph budget/massCheck info for all P fractions (units=Mg) */ 01355 sprintf( modelFileName, "%s/%s/Output/Budget/budg_P2", OutputPath, ProjName ); 01356 if ( ( budget_P2 = fopen( modelFileName, "w" ) ) == NULL ) 01357 {printf( "Can't open budg_P2 file! " );exit(-1) ;} 01358 /* Phosph budget/massCheck info for all P fractions (units=mg/m2) */ 01359 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Par2", OutputPath, ProjName ); 01360 if ( ( budget_Par2 = fopen( modelFileName, "w" ) ) == NULL ) 01361 {printf( "Can't open budg_Par2 file! " );exit(-1) ;} 01362 /* Salt budget/massCheck info */ 01363 sprintf( modelFileName, "%s/%s/Output/Budget/budg_S2", OutputPath, ProjName ); 01364 if ( ( budget_S2 = fopen( modelFileName, "w" ) ) == NULL ) 01365 {printf( "Can't open budg_S2 file! " );exit(-1) ;} 01366 01367 if (!ESPmodeON) { 01368 /* Phosph budget/massCheck info for live P fraction */ 01369 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pliv2", OutputPath, ProjName ); 01370 if ( ( budget_Plive2 = fopen( modelFileName, "w" ) ) == NULL ) 01371 {printf( "Can't open budg_Pliv2 file! " );exit(-1) ;} 01372 /* Phosph budget/massCheck info for dead P fraction */ 01373 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pded2", OutputPath, ProjName ); 01374 if ( ( budget_Pdead2 = fopen( modelFileName, "w" ) ) == NULL ) 01375 {printf( "Can't open budg_Pded2 file! " );exit(-1) ;} 01376 /* Phosph budget/massCheck info for water-borne P fraction */ 01377 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pwat2", OutputPath, ProjName ); 01378 if ( ( budget_Pwat2 = fopen( modelFileName, "w" ) ) == NULL ) 01379 {printf( "Can't open budg_Pwat2 file! " );exit(-1) ;} 01380 }/* end of !ESPmodeON */ 01381 01382 } /* end of opening secondary files */ 01383 01384 01385 if (numBasn>=26) { 01386 /*Open the tertiary files for all of the budgets if we have more than 25 basins */ 01387 /* Indicator region avgs */ 01388 sprintf( modelFileName, "%s/%s/Output/Budget/BIRavg3", OutputPath, ProjName ); 01389 if ( ( BIRavg3 = fopen( modelFileName, "w" ) ) == NULL ) 01390 { printf( "Can't open BIRavg3 file! " ); exit(-1) ;} 01391 /* water budget/massCheck info - units in acre-feet */ 01392 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wacr3", OutputPath, ProjName ); 01393 if ( ( budget_Wacr3 = fopen( modelFileName, "w" ) ) == NULL ) 01394 { printf( "Can't open budg_Wacr3 file! " ); exit(-1) ;} 01395 /* water budget/massCheck info - units in cm height */ 01396 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wcm3", OutputPath, ProjName ); 01397 if ( ( budget_Wcm3 = fopen( modelFileName, "w" ) ) == NULL ) 01398 {printf( "Can't open budg_Wcm3 file! " ); exit(-1) ;} 01399 /* Phosph budget/massCheck info for all P fractions (units=Mg) */ 01400 sprintf( modelFileName, "%s/%s/Output/Budget/budg_P3", OutputPath, ProjName ); 01401 if ( ( budget_P3 = fopen( modelFileName, "w" ) ) == NULL ) 01402 {printf( "Can't open budg_P3 file! " );exit(-1) ;} 01403 /* Phosph budget/massCheck info for all P fractions (units=mg/m2) */ 01404 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Par3", OutputPath, ProjName ); 01405 if ( ( budget_Par3 = fopen( modelFileName, "w" ) ) == NULL ) 01406 {printf( "Can't open budg_Par3 file! " );exit(-1) ;} 01407 /* Salt budget/massCheck info */ 01408 sprintf( modelFileName, "%s/%s/Output/Budget/budg_S3", OutputPath, ProjName ); 01409 if ( ( budget_S3 = fopen( modelFileName, "w" ) ) == NULL ) 01410 {printf( "Can't open budg_S3 file! " );exit(-1) ;} 01411 01412 if (!ESPmodeON) { 01413 /* Phosph budget/massCheck info for live P fraction */ 01414 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pliv3", OutputPath, ProjName ); 01415 if ( ( budget_Plive3 = fopen( modelFileName, "w" ) ) == NULL ) 01416 {printf( "Can't open budg_Pliv3 file! " );exit(-1) ;} 01417 /* Phosph budget/massCheck info for dead P fraction */ 01418 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pded3", OutputPath, ProjName ); 01419 if ( ( budget_Pdead3 = fopen( modelFileName, "w" ) ) == NULL ) 01420 {printf( "Can't open budg_Pded3 file! " );exit(-1) ;} 01421 /* Phosph budget/massCheck info for water-borne P fraction */ 01422 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pwat3", OutputPath, ProjName ); 01423 if ( ( budget_Pwat3 = fopen( modelFileName, "w" ) ) == NULL ) 01424 {printf( "Can't open budg_Pwat3 file! " );exit(-1) ;} 01425 }/* end of !ESPmodeON */ 01426 01427 } /* end of opening tertiary files */ 01428 01429 if (numBasn>=39) { 01430 /*Open the fourth set of files for all of the budgets if we have more than 38 basins */ 01431 /* Indicator region avgs */ 01432 sprintf( modelFileName, "%s/%s/Output/Budget/BIRavg4", OutputPath, ProjName ); 01433 if ( ( BIRavg4 = fopen( modelFileName, "w" ) ) == NULL ) 01434 { printf( "Can't open BIRavg4 file! " ); exit(-1) ;} 01435 /* water budget/massCheck info - units in acre-feet */ 01436 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wacr4", OutputPath, ProjName ); 01437 if ( ( budget_Wacr4 = fopen( modelFileName, "w" ) ) == NULL ) 01438 { printf( "Can't open budg_Wacr4 file! " ); exit(-1) ;} 01439 /* water budget/massCheck info - units in cm height */ 01440 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wcm4", OutputPath, ProjName ); 01441 if ( ( budget_Wcm4 = fopen( modelFileName, "w" ) ) == NULL ) 01442 {printf( "Can't open budg_Wcm4 file! " ); exit(-1) ;} 01443 /* Phosph budget/massCheck info for all P fractions (units=Mg) */ 01444 sprintf( modelFileName, "%s/%s/Output/Budget/budg_P4", OutputPath, ProjName ); 01445 if ( ( budget_P4 = fopen( modelFileName, "w" ) ) == NULL ) 01446 {printf( "Can't open budg_P4 file! " );exit(-1) ;} 01447 /* Phosph budget/massCheck info for all P fractions (units=mg/m2) */ 01448 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Par4", OutputPath, ProjName ); 01449 if ( ( budget_Par4 = fopen( modelFileName, "w" ) ) == NULL ) 01450 {printf( "Can't open budg_Par4 file! " );exit(-1) ;} 01451 /* Salt budget/massCheck info */ 01452 sprintf( modelFileName, "%s/%s/Output/Budget/budg_S4", OutputPath, ProjName ); 01453 if ( ( budget_S4 = fopen( modelFileName, "w" ) ) == NULL ) 01454 {printf( "Can't open budg_S4 file! " );exit(-1) ;} 01455 01456 if (!ESPmodeON) { 01457 /* Phosph budget/massCheck info for live P fraction */ 01458 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pliv4", OutputPath, ProjName ); 01459 if ( ( budget_Plive4 = fopen( modelFileName, "w" ) ) == NULL ) 01460 {printf( "Can't open budg_Pliv4 file! " );exit(-1) ;} 01461 /* Phosph budget/massCheck info for dead P fraction */ 01462 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pded4", OutputPath, ProjName ); 01463 if ( ( budget_Pdead4 = fopen( modelFileName, "w" ) ) == NULL ) 01464 {printf( "Can't open budg_Pded4 file! " );exit(-1) ;} 01465 /* Phosph budget/massCheck info for water-borne P fraction */ 01466 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pwat4", OutputPath, ProjName ); 01467 if ( ( budget_Pwat4 = fopen( modelFileName, "w" ) ) == NULL ) 01468 {printf( "Can't open budg_Pwat4 file! " );exit(-1) ;} 01469 }/* end of !ESPmodeON */ 01470 01471 } /* end of opening fourth set of files */ 01472 01473 if (numBasn>=52) { 01474 /*Open the fifth set of files for all of the budgets if we have more than 51 basins */ 01475 /* Indicator region avgs */ 01476 sprintf( modelFileName, "%s/%s/Output/Budget/BIRavg5", OutputPath, ProjName ); 01477 if ( ( BIRavg5 = fopen( modelFileName, "w" ) ) == NULL ) 01478 { printf( "Can't open BIRavg5 file! " ); exit(-1) ;} 01479 /* water budget/massCheck info - units in acre-feet */ 01480 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wacr5", OutputPath, ProjName ); 01481 if ( ( budget_Wacr5 = fopen( modelFileName, "w" ) ) == NULL ) 01482 { printf( "Can't open budg_Wacr5 file! " ); exit(-1) ;} 01483 /* water budget/massCheck info - units in cm height */ 01484 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Wcm5", OutputPath, ProjName ); 01485 if ( ( budget_Wcm5 = fopen( modelFileName, "w" ) ) == NULL ) 01486 {printf( "Can't open budg_Wcm5 file! " ); exit(-1) ;} 01487 /* Phosph budget/massCheck info for all P fractions (units=Mg) */ 01488 sprintf( modelFileName, "%s/%s/Output/Budget/budg_P5", OutputPath, ProjName ); 01489 if ( ( budget_P5 = fopen( modelFileName, "w" ) ) == NULL ) 01490 {printf( "Can't open budg_P5 file! " );exit(-1) ;} 01491 /* Phosph budget/massCheck info for all P fractions (units=mg/m2) */ 01492 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Par5", OutputPath, ProjName ); 01493 if ( ( budget_Par5 = fopen( modelFileName, "w" ) ) == NULL ) 01494 {printf( "Can't open budg_Par5 file! " );exit(-1) ;} 01495 /* Salt budget/massCheck info */ 01496 sprintf( modelFileName, "%s/%s/Output/Budget/budg_S5", OutputPath, ProjName ); 01497 if ( ( budget_S5 = fopen( modelFileName, "w" ) ) == NULL ) 01498 {printf( "Can't open budg_S5 file! " );exit(-1) ;} 01499 01500 if (!ESPmodeON) { 01501 /* Phosph budget/massCheck info for live P fraction */ 01502 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pliv5", OutputPath, ProjName ); 01503 if ( ( budget_Plive5 = fopen( modelFileName, "w" ) ) == NULL ) 01504 {printf( "Can't open budg_Pliv5 file! " );exit(-1) ;} 01505 /* Phosph budget/massCheck info for dead P fraction */ 01506 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pded5", OutputPath, ProjName ); 01507 if ( ( budget_Pdead5 = fopen( modelFileName, "w" ) ) == NULL ) 01508 {printf( "Can't open budg_Pded5 file! " );exit(-1) ;} 01509 /* Phosph budget/massCheck info for water-borne P fraction */ 01510 sprintf( modelFileName, "%s/%s/Output/Budget/budg_Pwat5", OutputPath, ProjName ); 01511 if ( ( budget_Pwat5 = fopen( modelFileName, "w" ) ) == NULL ) 01512 {printf( "Can't open budg_Pwat5 file! " );exit(-1) ;} 01513 }/* end of !ESPmodeON */ 01514 01515 } /* end of opening fifth set of files */ 01516 01517 } /* end of opening all files */ 01518 01519 01520 01521 /********/ 01522 /* Now we need to print the headers to each set(s) of basin files */ 01523 /* First, figure out the number of files sets, and then loop thru that */ 01524 /* there are 13 total basins printed per file (basin 0 is whole system, 01525 not counted in numBasn, but always printed out in first file) */ 01526 if (numBasn>=52) { /* with more than 58 (plus whole-system basin == 59) basins: */ 01527 Fsets=5; /* we have five sets of basin files */ 01528 } 01529 else if (numBasn>=39) { /* with more than 38 (plus whole-system basin == 39) basins: */ 01530 Fsets=4; /* we have four sets of basin files */ 01531 } 01532 else if (numBasn>=26) { /* with more than 25 (plus whole-system basin == 26) basins: */ 01533 Fsets=3; /* we have three sets of basin files */ 01534 } 01535 else if (numBasn>=13) { /* with more than 12 (plus whole-system basin == 13) basins: */ 01536 Fsets=2; /* we have two sets of basin files */ 01537 } 01538 else { 01539 Fsets=1; /* otherwise, just one set of files */ 01540 } 01541 01542 for (Fnum = 1; Fnum <= Fsets; Fnum++) { 01543 01544 if (Fnum == 1) { /* first file set */ 01545 if (Fsets > 1) { 01546 ibasLow = 0; 01547 ibasHigh = 12; 01548 } 01549 else { 01550 ibasLow = 0; 01551 ibasHigh = Min(12,numBasn); 01552 } 01553 } 01554 01555 if (Fnum == 2) { /* second file set */ 01556 if (Fsets > 2) { 01557 ibasLow = 13; 01558 ibasHigh = 25; 01559 } 01560 else { 01561 ibasLow = 13; 01562 ibasHigh = Min(25,numBasn); 01563 } 01564 } 01565 01566 if (Fnum == 3) { /* third file set */ 01567 if (Fsets > 3) { 01568 ibasLow = 26; 01569 ibasHigh = 38; 01570 } 01571 else { 01572 ibasLow = 26; 01573 ibasHigh = Min(38,numBasn); 01574 } 01575 } 01576 01577 if (Fnum == 4) { /* fourth file set */ 01578 if (Fsets > 4) { 01579 ibasLow = 39; 01580 ibasHigh = 51; 01581 } 01582 else { 01583 ibasLow = 39; 01584 ibasHigh = Min(51,numBasn); 01585 } 01586 } 01587 01588 01589 else if (Fnum == 5) { 01590 ibasLow = 52; 01591 ibasHigh = numBasn; 01592 } 01593 01594 /*****/ 01595 /* now print the 4 header lines for all of the budget & BIRavg files */ 01596 01597 /* LINE 1&2: first header line, providing scenario ID; second line is units, etc. */ 01598 /* feb05 added two tabs at end of BIRavg header to accomodate sensitivity parm label & value */ 01599 fprintf ( BIRhydro, 01600 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): daily mean attributes. \nUnits = Stage_minus_LandElev (StgMinElev) == m, Surface_Velocity (BIR_Sf_vel) = m/d, DayFire == days, CLsf_avg == CL conc. in sfwat, g/L.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01601 01602 fprintf ( ((Fnum==5)?(BIRavg5): 01603 ((Fnum==4)?(BIRavg4): 01604 ((Fnum==3)?(BIRavg3): 01605 ((Fnum==2)?(BIRavg2): 01606 (BIRavg1) ) ) ) ), 01607 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): daily mean attributes. \nUnits = Hydro depths== m, TP surface & pore water==mg/L, TP soil==mg/kg, noncalc&calc periph==gC/m2, mac==kgC/m2, LandElevation= m NGVD/NAVD+DATUM_DISTANCE(usually 6m).\n \t\t\t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01608 fprintf ( ((Fnum==5)?(budget_Wacr5): 01609 ((Fnum==4)?(budget_Wacr4): 01610 ((Fnum==3)?(budget_Wacr3): 01611 ((Fnum==2)?(budget_Wacr2): 01612 (budget_Wacr1) ) ) ) ), 01613 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): water budget and mass balance check. \nUnits = thousands of acre-feet per basin, with error (and cumulative net error) reporting in mm height across basin.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01614 fprintf ( ((Fnum==5)?(budget_Wcm5): 01615 ((Fnum==4)?(budget_Wcm4): 01616 ((Fnum==3)?(budget_Wcm3): 01617 ((Fnum==2)?(budget_Wcm2): 01618 (budget_Wcm1) ) ) ) ), 01619 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): water budget and mass balance check. \nUnits = cm per basin, with error (and cumulative net error) reporting in mm height across basin.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01620 fprintf ( ((Fnum==5)?(budget_S5): 01621 ((Fnum==4)?(budget_S4): 01622 ((Fnum==3)?(budget_S3): 01623 ((Fnum==2)?(budget_S2): 01624 (budget_S1) ) ) ) ), 01625 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): conservative tracer budget and mass balance check. \nUnits = metric tons (Mg) per basin, with error (and cumulative net error) reporting in ug/m2 in basin.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01626 fprintf ( ((Fnum==5)?(budget_P5): 01627 ((Fnum==4)?(budget_P4): 01628 ((Fnum==3)?(budget_P3): 01629 ((Fnum==2)?(budget_P2): 01630 (budget_P1) ) ) ) ), 01631 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): Total P budget and mass balance check for all fractions. \nUnits = metric tons (Mg) per basin, with error (and cumulative net error) reporting in ug/m2 in basin.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01632 fprintf ( ((Fnum==5)?(budget_Par5): 01633 ((Fnum==4)?(budget_Par4): 01634 ((Fnum==3)?(budget_Par3): 01635 ((Fnum==2)?(budget_Par2): 01636 (budget_Par1) ) ) ) ), 01637 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): Total P budget and mass balance check for all fractions. \nUnits = mg/m2 in basin, with error (and cumulative net error) reporting in ug/m2 in basin.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01638 if (!ESPmodeON) { 01639 01640 fprintf ( ((Fnum==5)?(budget_Plive5): 01641 ((Fnum==4)?(budget_Plive4): 01642 ((Fnum==3)?(budget_Plive3): 01643 ((Fnum==2)?(budget_Plive2): 01644 (budget_Plive1) ) ) ) ), 01645 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): Total P budget and mass balance check for live fraction. \nUnits = mg/m2 in basin, with error (and cumulative net error) ug/m2.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01646 fprintf ( ((Fnum==5)?(budget_Pdead5): 01647 ((Fnum==4)?(budget_Pdead4): 01648 ((Fnum==3)?(budget_Pdead3): 01649 ((Fnum==2)?(budget_Pdead2): 01650 (budget_Pdead1) ) ) ) ), 01651 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): Total P budget and mass balance check for dead, non-water-borne fraction. \nUnits = mg/m2 in basin, with error (and cumulative net error) ug/m2.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01652 fprintf ( ((Fnum==5)?(budget_Pwat5): 01653 ((Fnum==4)?(budget_Pwat4): 01654 ((Fnum==3)?(budget_Pwat3): 01655 ((Fnum==2)?(budget_Pwat2): 01656 (budget_Pwat1) ) ) ) ), 01657 "%s %s %s %s scenario: Basins/Indicator-Regions (BIR): Total P budget and mass balance check for water-borne fraction (some horiz I/O not shown - see budget for all combined fractions). \nUnits = mg/m2 in basin, with error (and cumulative net error) ug/m2.\n \t ", &modelName, &modelVers, &SimAlt, &SimModif ); 01658 } 01659 01660 /* LINE 3: third header line, looping thru basins, providing basin ID and size */ 01661 for (ibas = ibasHigh; ibas >= ibasLow; ibas--) 01662 { 01663 /* v2.8 hydro Perf Measure summaries - is a first cut, TODO is re-organize it */ 01664 fprintf ( BIRhydro, 01665 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t", 01666 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01667 01668 fprintf ( ((Fnum==5)?(BIRavg5): 01669 ((Fnum==4)?(BIRavg4): 01670 ((Fnum==3)?(BIRavg3): 01671 ((Fnum==2)?(BIRavg2): 01672 (BIRavg1) ) ) ) ), 01673 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t", 01674 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01675 fprintf ( ((Fnum==5)?(budget_Wacr5): 01676 ((Fnum==4)?(budget_Wacr4): 01677 ((Fnum==3)?(budget_Wacr3): 01678 ((Fnum==2)?(budget_Wacr2): 01679 (budget_Wacr1) ) ) ) ), 01680 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t \t \t \t \t", 01681 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01682 fprintf ( ((Fnum==5)?(budget_Wcm5): 01683 ((Fnum==4)?(budget_Wcm4): 01684 ((Fnum==3)?(budget_Wcm3): 01685 ((Fnum==2)?(budget_Wcm2): 01686 (budget_Wcm1) ) ) ) ), 01687 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t \t \t \t \t", 01688 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01689 fprintf ( ((Fnum==5)?(budget_S5): 01690 ((Fnum==4)?(budget_S4): 01691 ((Fnum==3)?(budget_S3): 01692 ((Fnum==2)?(budget_S2): 01693 (budget_S1) ) ) ) ), 01694 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t \t", 01695 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01696 if (!ESPmodeON) { 01697 fprintf ( ((Fnum==5)?(budget_P5): 01698 ((Fnum==4)?(budget_P4): 01699 ((Fnum==3)?(budget_P3): 01700 ((Fnum==2)?(budget_P2): 01701 (budget_P1) ) ) ) ), 01702 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t \t", 01703 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01704 fprintf ( ((Fnum==5)?(budget_Par5): 01705 ((Fnum==4)?(budget_Par4): 01706 ((Fnum==3)?(budget_Par3): 01707 ((Fnum==2)?(budget_Par2): 01708 (budget_Par1) ) ) ) ), 01709 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t \t", 01710 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01711 fprintf ( ((Fnum==5)?(budget_Plive5): 01712 ((Fnum==4)?(budget_Plive4): 01713 ((Fnum==3)?(budget_Plive3): 01714 ((Fnum==2)?(budget_Plive2): 01715 (budget_Plive1) ) ) ) ), 01716 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t", 01717 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01718 fprintf ( ((Fnum==5)?(budget_Pdead5): 01719 ((Fnum==4)?(budget_Pdead4): 01720 ((Fnum==3)?(budget_Pdead3): 01721 ((Fnum==2)?(budget_Pdead2): 01722 (budget_Pdead1) ) ) ) ), 01723 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t", 01724 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01725 fprintf ( ((Fnum==5)?(budget_Pwat5): 01726 ((Fnum==4)?(budget_Pwat4): 01727 ((Fnum==3)?(budget_Pwat3): 01728 ((Fnum==2)?(budget_Pwat2): 01729 (budget_Pwat1) ) ) ) ), 01730 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t \t \t", 01731 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01732 } 01733 01734 else { 01735 fprintf ( ((Fnum==5)?(budget_P5): 01736 ((Fnum==4)?(budget_P4): 01737 ((Fnum==3)?(budget_P3): 01738 ((Fnum==2)?(budget_P2): 01739 (budget_P1) ) ) ) ), 01740 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t \t \t", 01741 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01742 fprintf ( ((Fnum==5)?(budget_Par5): 01743 ((Fnum==4)?(budget_Par4): 01744 ((Fnum==3)?(budget_Par3): 01745 ((Fnum==2)?(budget_Par2): 01746 (budget_Par1) ) ) ) ), 01747 " BIR %2d =\t%6.1fmi^2,\t%6.1fkm^2\t \t \t \t \t \t \t \t \t \t \t \t \t \t \t", 01748 ibas,(numCells[ibas]*CELL_SIZE*3.8610039e-7),(numCells[ibas]*CELL_SIZE*1.0e-6) ); 01749 } 01750 } /*end of loop for second header line */ 01751 01752 /* LINE 4: beginning of fourth header line */ 01753 /* feb05 added two tabs prior to "Date" of BIRavg header to accomodate sensitivity parm label & value */ 01754 fprintf ( BIRhydro, "\nDate\t" ); 01755 01756 fprintf ( ((Fnum==5)?(BIRavg5): 01757 ((Fnum==4)?(BIRavg4): 01758 ((Fnum==3)?(BIRavg3): 01759 ((Fnum==2)?(BIRavg2): 01760 (BIRavg1) ) ) ) ), "\nSParm\tSParmValu\t Date\t" ); 01761 fprintf ( ((Fnum==5)?(budget_Wacr5): 01762 ((Fnum==4)?(budget_Wacr4): 01763 ((Fnum==3)?(budget_Wacr3): 01764 ((Fnum==2)?(budget_Wacr2): 01765 (budget_Wacr1) ) ) ) ), "\n Date\t" ); 01766 fprintf ( ((Fnum==5)?(budget_Wcm5): 01767 ((Fnum==4)?(budget_Wcm4): 01768 ((Fnum==3)?(budget_Wcm3): 01769 ((Fnum==2)?(budget_Wcm2): 01770 (budget_Wcm1) ) ) ) ), "\n Date\t" ); 01771 fprintf ( ((Fnum==5)?(budget_S5): 01772 ((Fnum==4)?(budget_S4): 01773 ((Fnum==3)?(budget_S3): 01774 ((Fnum==2)?(budget_S2): 01775 (budget_S1) ) ) ) ), "\n Date\t" ); 01776 fprintf ( ((Fnum==5)?(budget_P5): 01777 ((Fnum==4)?(budget_P4): 01778 ((Fnum==3)?(budget_P3): 01779 ((Fnum==2)?(budget_P2): 01780 (budget_P1) ) ) ) ), "\n Date\t" ); 01781 fprintf ( ((Fnum==5)?(budget_Par5): 01782 ((Fnum==4)?(budget_Par4): 01783 ((Fnum==3)?(budget_Par3): 01784 ((Fnum==2)?(budget_Par2): 01785 (budget_Par1) ) ) ) ), "\n Date\t" ); 01786 if (!ESPmodeON) { 01787 fprintf ( ((Fnum==5)?(budget_Plive5): 01788 ((Fnum==4)?(budget_Plive4): 01789 ((Fnum==3)?(budget_Plive3): 01790 ((Fnum==2)?(budget_Plive2): 01791 (budget_Plive1) ) ) ) ), "\n Date\t" ); 01792 fprintf ( ((Fnum==5)?(budget_Pdead5): 01793 ((Fnum==4)?(budget_Pdead4): 01794 ((Fnum==3)?(budget_Pdead3): 01795 ((Fnum==2)?(budget_Pdead2): 01796 (budget_Pdead1) ) ) ) ), "\n Date\t" ); 01797 fprintf ( ((Fnum==5)?(budget_Pwat5): 01798 ((Fnum==4)?(budget_Pwat4): 01799 ((Fnum==3)?(budget_Pwat3): 01800 ((Fnum==2)?(budget_Pwat2): 01801 (budget_Pwat1) ) ) ) ), "\n Date\t" ); 01802 } 01803 01804 /* LINE 4: remainder of fourth header line, looping thru basins, providing column/variable ID attributes */ 01805 for (ibas = ibasHigh; ibas >= ibasLow; ibas--) 01806 { 01807 01808 fprintf ( BIRhydro, 01809 " StgMinElev_%d\t Veloc_%d\t DayFire_%d\t CLsf_avg_%d\t", 01810 ibas,ibas,ibas,ibas); 01811 01812 fprintf ( ((Fnum==5)?(BIRavg5): 01813 ((Fnum==4)?(BIRavg4): 01814 ((Fnum==3)?(BIRavg3): 01815 ((Fnum==2)?(BIRavg2): 01816 (BIRavg1) ) ) ) ), 01817 " SfWat_%d\t Unsat_%d\t TPsf_%d\t TPpore_%d\t TPsoil_%d\t NCperi_%d\t Cperi_%d\t Mac_%d\t Elev_%d\t", 01818 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01819 fprintf ( ((Fnum==5)?(budget_Wacr5): 01820 ((Fnum==4)?(budget_Wacr4): 01821 ((Fnum==3)?(budget_Wacr3): 01822 ((Fnum==2)?(budget_Wacr2): 01823 (budget_Wacr1) ) ) ) ), 01824 " VolOld_%d\t Atmos_%d\t Evap_%d\t Transp_%d\t Rchg_%d\t StrIn_%d\t StrOut_%d\t OvlIn_%d\t OvlOut_%d\t SpgIn_%d\t SpgOut_%d\t GWin_%d\t GWout_%d\t VolTarg_%d\t VolNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01825 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01826 fprintf ( ((Fnum==5)?(budget_Wcm5): 01827 ((Fnum==4)?(budget_Wcm4): 01828 ((Fnum==3)?(budget_Wcm3): 01829 ((Fnum==2)?(budget_Wcm2): 01830 (budget_Wcm1) ) ) ) ), 01831 " VolOld_%d\t Atmos_%d\t Evap_%d\t Transp_%d\t Rchg_%d\t StrIn_%d\t StrOut_%d\t OvlIn_%d\t OvlOut_%d\t SpgIn_%d\t SpgOut_%d\t GWin_%d\t GWout_%d\t VolTarg_%d\t VolNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01832 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01833 fprintf ( ((Fnum==5)?(budget_S5): 01834 ((Fnum==4)?(budget_S4): 01835 ((Fnum==3)?(budget_S3): 01836 ((Fnum==2)?(budget_S2): 01837 (budget_S1) ) ) ) ), 01838 " MasOld_%d\t Atmos_%d\t StrIn_%d\t StrOut_%d\t OvlIn_%d\t OvlOut_%d\t SpgIn_%d\t SpgOut_%d\t GWin_%d\t GWout_%d\t MasTarg_%d\t MasNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01839 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01840 if (!ESPmodeON) { 01841 fprintf ( ((Fnum==5)?(budget_P5): 01842 ((Fnum==4)?(budget_P4): 01843 ((Fnum==3)?(budget_P3): 01844 ((Fnum==2)?(budget_P2): 01845 (budget_P1) ) ) ) ), 01846 " MasOld_%d\t Atmos_%d\t StrIn_%d\t StrOut_%d\t OvlIn_%d\t OvlOut_%d\t SpgIn_%d\t SpgOut_%d\t GWin_%d\t GWout_%d\t MasTarg_%d\t MasNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01847 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01848 fprintf ( ((Fnum==5)?(budget_Par5): 01849 ((Fnum==4)?(budget_Par4): 01850 ((Fnum==3)?(budget_Par3): 01851 ((Fnum==2)?(budget_Par2): 01852 (budget_Par1) ) ) ) ), 01853 " MasOld_%d\t Atmos_%d\t StrIn_%d\t StrOut_%d\t OvlIn_%d\t OvlOut_%d\t SpgIn_%d\t SpgOut_%d\t GWin_%d\t GWout_%d\t MasTarg_%d\t MasNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01854 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01855 fprintf ( ((Fnum==5)?(budget_Plive5): 01856 ((Fnum==4)?(budget_Plive4): 01857 ((Fnum==3)?(budget_Plive3): 01858 ((Fnum==2)?(budget_Plive2): 01859 (budget_Plive1) ) ) ) ), 01860 " MacBio_%d\t CPerBio_%d\t NCPerBio_%d\t MacNPP_%d\t CPerGPP_%d\tNCPerGPP_%d\t MacMort_%d\t CalgMort_%d\t NCalgMort_%d\t MasTarg_%d\t MasNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01861 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01862 fprintf ( ((Fnum==5)?(budget_Pdead5): 01863 ((Fnum==4)?(budget_Pdead4): 01864 ((Fnum==3)?(budget_Pdead3): 01865 ((Fnum==2)?(budget_Pdead2): 01866 (budget_Pdead1) ) ) ) ), 01867 " MasOld_%d\tMac-soil_%d\tPer-soil_%d\t Decomp_%d\tSettl_to_%d\t Sorb_to_%d\t Desorb_%d\t MasTarg_%d\t MasNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01868 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01869 fprintf ( ((Fnum==5)?(budget_Pwat5): 01870 ((Fnum==4)?(budget_Pwat4): 01871 ((Fnum==3)?(budget_Pwat3): 01872 ((Fnum==2)?(budget_Pwat2): 01873 (budget_Pwat1) ) ) ) ), 01874 " MasOld_%d\t Atmos_%d\tSettl_fr_%d\t SfMiner_%d\tSedMiner_%d\t AlgUpt_%d\t MacUpt_%d\t DeSorb_%d\t Sorb_to_%d\t StrIn_%d\t StrOut_%d\t MasTarg_%d\t MasNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01875 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01876 } 01877 else { 01878 fprintf ( ((Fnum==5)?(budget_P5): 01879 ((Fnum==4)?(budget_P4): 01880 ((Fnum==3)?(budget_P3): 01881 ((Fnum==2)?(budget_P2): 01882 (budget_P1) ) ) ) ), 01883 " MasOld_%d\t Atmos_%d\t Settl_%d\t StrIn_%d\t StrOut_%d\t OvlIn_%d\t OvlOut_%d\t SpgIn_%d\t SpgOut_%d\t GWin_%d\t GWout_%d\t MasTarg_%d\t MasNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01884 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01885 fprintf ( ((Fnum==5)?(budget_Par5): 01886 ((Fnum==4)?(budget_Par4): 01887 ((Fnum==3)?(budget_Par3): 01888 ((Fnum==2)?(budget_Par2): 01889 (budget_Par1) ) ) ) ), 01890 " MasOld_%d\t Atmos_%d\t Settl_%d\t StrIn_%d\t StrOut_%d\t OvlIn_%d\t OvlOut_%d\t SpgIn_%d\t SpgOut_%d\t GWin_%d\t GWout_%d\t MasTarg_%d\t MasNew_%d\t Err_%d\t SumErr_%d\t InAvg_%d\t OutAvg_%d\t", 01891 ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas,ibas); 01892 } 01893 } /* end of loop for third header line */ 01894 01895 } /*end of printing all files' headers */ 01896 01897 01898 } /* end of BIRoutfiles */
| void BIRstats_reset | ( | void | ) |
Reset BIR (non-budget) statistics summations to zero.
- Parameters:
-
ibas Basin/Indicator-Region ID number
Definition at line 1028 of file BudgStats.c.
References BIRavg1, BIRavg2, BIRavg3, BIRavg4, BIRavg5, Cperi_avg, Elev_avg, Mac_avg, NCperi_avg, numBasn, Sfwat_avg, TPpore_avg, TPsf_avg, TPsoil_avg, and Unsat_avg.
01029 { 01030 int ibas; 01031 01032 for (ibas = numBasn; ibas >= 0; ibas--) { 01033 Sfwat_avg[ibas]=Unsat_avg[ibas]=TPsf_avg[ibas]=TPpore_avg[ibas]=TPsoil_avg[ibas]= 01034 NCperi_avg[ibas]=Cperi_avg[ibas]=Mac_avg[ibas]=Elev_avg[ibas]=0.0; 01035 } 01036 fflush (BIRavg1); 01037 if (numBasn>=13) fflush (BIRavg2); 01038 if (numBasn>=26) fflush (BIRavg3); 01039 if (numBasn>=39) fflush (BIRavg4); 01040 if (numBasn>=52) fflush (BIRavg5); 01041 01042 }
| void BIRbudg_reset | ( | void | ) |
Reset BIR budget summations to zero.
For ALL budgets, store the old total volume in array TOT_xxx_OLD and reset any summations and BIRavgs to 0.
- Parameters:
-
ibas Basin/Indicator-Region ID number
Definition at line 1066 of file BudgStats.c.
References budget_P1, budget_P2, budget_P3, budget_P4, budget_P5, budget_Par1, budget_Par2, budget_Par3, budget_Par4, budget_Par5, budget_Pdead1, budget_Pdead2, budget_Pdead3, budget_Pdead4, budget_Pdead5, budget_Plive1, budget_Plive2, budget_Plive3, budget_Plive4, budget_Plive5, budget_Pwat1, budget_Pwat2, budget_Pwat3, budget_Pwat4, budget_Pwat5, budget_S1, budget_S2, budget_S3, budget_S4, budget_S5, budget_Wacr1, budget_Wacr2, budget_Wacr3, budget_Wacr4, budget_Wacr5, budget_Wcm1, budget_Wcm2, budget_Wcm3, budget_Wcm4, budget_Wcm5, Calg_GPP, Calg_mort, dop_decomp, dop_desorb, dop_macIn, dop_sorbIn, ESPmodeON, EVAP, floc_decomp, floc_In, mac_mort, mac_NPP, NCalg_GPP, NCalg_mort, numBasn, P, P_ATMOS, P_Calg, P_CELL, P_DEAD, P_DEAD_CELL, P_DEAD_OLD, P_IN_GW, P_IN_OVL, P_IN_SPG, P_IN_STR, P_LIVE, P_LIVE_CELL, P_LIVE_OLD, P_MAC, P_NCalg, P_OLD, P_OUT_GW, P_OUT_OVL, P_OUT_SPG, P_OUT_STR, P_settl, P_WAT, P_WAT_CELL, P_WAT_OLD, RAIN, RCHG, S_IN_GW, S_IN_OVL, S_IN_SPG, S_IN_STR, S_OUT_GW, S_OUT_OVL, S_OUT_SPG, S_OUT_STR, SALT_ATMOS, SUMGW, SUMSF, SUMUW, TOT_P_CAN, TOT_S, TOT_S_CAN, TOT_S_CELL, TOT_S_OLD, TOT_VOL, TOT_VOL_CAN, TOT_VOL_OLD, TRANSP, VOL_IN_GW, VOL_IN_OVL, VOL_IN_SPG, VOL_IN_STR, VOL_OUT_GW, VOL_OUT_OVL, VOL_OUT_SPG, VOL_OUT_STR, wat_sedMiner, wat_sedUpt, wat_sfMiner, and wat_sfUpt.
01067 { 01068 int ibas; 01069 01070 for (ibas = numBasn; ibas >= 0; ibas--) { 01071 TOT_VOL_OLD[ibas] = TOT_VOL[ibas]; 01072 TOT_S_OLD[ibas] = TOT_S[ibas]; 01073 P_OLD[ibas] = P[ibas]; 01074 if (!ESPmodeON) { 01075 P_LIVE_OLD[ibas] = P_LIVE[ibas]; 01076 P_DEAD_OLD[ibas] = P_DEAD[ibas]; 01077 P_WAT_OLD[ibas] = P_WAT[ibas]; 01078 } 01079 01080 SUMSF[ibas] = SUMGW[ibas] = SUMUW[ibas] = TOT_VOL[ibas] = TOT_VOL_CAN[ibas] = 0.0; 01081 RAIN[ibas] = VOL_IN_STR[ibas] = VOL_IN_OVL[ibas] = VOL_IN_SPG[ibas] = VOL_IN_GW[ibas] = 0.0; 01082 EVAP[ibas] = TRANSP[ibas] = RCHG[ibas] = VOL_OUT_STR[ibas] = VOL_OUT_OVL[ibas] = VOL_OUT_SPG[ibas] = VOL_OUT_GW[ibas] = 0.0; 01083 01084 TOT_S[ibas] = TOT_S_CELL[ibas] = TOT_S_CAN[ibas] = 0.0; 01085 SALT_ATMOS[ibas] = S_IN_STR[ibas] = S_IN_OVL[ibas] = S_IN_SPG[ibas] = S_IN_GW[ibas] = 0.0; 01086 S_OUT_STR[ibas] = S_OUT_OVL[ibas] = S_OUT_SPG[ibas] = S_OUT_GW[ibas] = 0.0; 01087 01088 P[ibas] = P_CELL[ibas] = TOT_P_CAN[ibas] = 0.0; 01089 P_ATMOS[ibas] = P_IN_STR[ibas] = P_IN_OVL[ibas] = P_IN_SPG[ibas] = P_IN_GW[ibas] = 0.0; 01090 P_OUT_STR[ibas] = P_OUT_OVL[ibas] = P_OUT_SPG[ibas] = P_OUT_GW[ibas] = 0.0; 01091 01092 P_LIVE[ibas] = P_LIVE_CELL[ibas] = P_MAC[ibas] = P_Calg[ibas] = P_NCalg[ibas] = 0.0; 01093 Calg_GPP[ibas] = NCalg_GPP[ibas] = Calg_mort[ibas] = NCalg_mort[ibas] = 0.0; 01094 mac_NPP[ibas] = mac_mort[ibas] = 0.0; 01095 01096 P_DEAD[ibas] = P_DEAD_CELL[ibas] = 0.0; 01097 dop_macIn[ibas] = dop_sorbIn[ibas] = floc_In[ibas] = 0.0; 01098 dop_decomp[ibas] = dop_desorb[ibas] = floc_decomp[ibas] = 0.0; 01099 01100 P_WAT[ibas] = P_WAT_CELL[ibas] = 0.0; 01101 wat_sfMiner[ibas] = wat_sedMiner[ibas] = wat_sfUpt[ibas] = wat_sedUpt[ibas] = P_settl[ibas] = 0.0; 01102 01103 01104 } 01105 01106 01107 fflush (budget_Wacr1); 01108 if (numBasn>=13) fflush (budget_Wacr2); 01109 if (numBasn>=26) fflush (budget_Wacr3); 01110 if (numBasn>=39) fflush (budget_Wacr4); 01111 if (numBasn>=52) fflush (budget_Wacr5); 01112 01113 fflush (budget_Wcm1); 01114 if (numBasn>=13) fflush (budget_Wcm2); 01115 if (numBasn>=26) fflush (budget_Wcm3); 01116 if (numBasn>=39) fflush (budget_Wcm4); 01117 if (numBasn>=52) fflush (budget_Wcm5); 01118 01119 fflush (budget_S1); 01120 if (numBasn>=13) fflush (budget_S2); 01121 if (numBasn>=26) fflush (budget_S3); 01122 if (numBasn>=39) fflush (budget_S4); 01123 if (numBasn>=52) fflush (budget_S5); 01124 01125 fflush (budget_P1); 01126 if (numBasn>=13) fflush (budget_P2); 01127 if (numBasn>=26) fflush (budget_P3); 01128 if (numBasn>=39) fflush (budget_P4); 01129 if (numBasn>=52) fflush (budget_P5); 01130 01131 fflush (budget_Par1); 01132 if (numBasn>=13) fflush (budget_Par2); 01133 if (numBasn>=26) fflush (budget_Par3); 01134 if (numBasn>=39) fflush (budget_Par4); 01135 if (numBasn>=52) fflush (budget_Par5); 01136 01137 if (!ESPmodeON) { 01138 fflush (budget_Plive1); 01139 if (numBasn>=13) fflush (budget_Plive2); 01140 if (numBasn>=26) fflush (budget_Plive3); 01141 if (numBasn>=39) fflush (budget_Plive4); 01142 if (numBasn>=52) fflush (budget_Plive5); 01143 01144 fflush (budget_Pdead1); 01145 if (numBasn>=13) fflush (budget_Pdead2); 01146 if (numBasn>=26) fflush (budget_Pdead3); 01147 if (numBasn>=39) fflush (budget_Pdead4); 01148 if (numBasn>=52) fflush (budget_Pdead5); 01149 01150 fflush (budget_Pwat1); 01151 if (numBasn>=13) fflush (budget_Pwat2); 01152 if (numBasn>=26) fflush (budget_Pwat3); 01153 if (numBasn>=39) fflush (budget_Pwat4); 01154 if (numBasn>=52) fflush (budget_Pwat5); 01155 } 01156 } /* end of BIRbudg_reset() */
| void Cell_reset_avg | ( | void | ) |
Zero the arrays holding selected variable averages in cells (after printing)/.
Definition at line 2065 of file BudgStats.c.
References BCmodel_sfwatAvg, C_Peri_mortAvg, C_Peri_nppAvg, C_Peri_PCAvg, C_PeriAvg, C_PeriNutCFAvg, C_PeriRespAvg, ETAvg, EvapAvg, Floc_fr_phBioAvg, HydRelDepPosNegAvg, LAI_effAvg, mac_nph_PCAvg, Mac_nphBioAvg, Mac_nphMortAvg, Mac_nppAvg, mac_ph_PCAvg, Mac_phBioAvg, Mac_phMortAvg, Mac_totBioAvg, MacNutCfAvg, MacWatCfAvg, Manning_nAvg, NC_Peri_mortAvg, NC_Peri_nppAvg, NC_PeriAvg, NC_PeriNutCFAvg, NC_PeriRespAvg, ON_MAP, PeriAvg, PeriLiteCFAvg, RainAvg, s0, s1, SaltSedAvg, SaltSfAvg, SedElevAvg, SF_WT_VEL_magAvg, SfWatAvg, T, TotHeadAvg, TP_settlAvg, TPSedMinAvg, TPSedUptAvg, TPSedWatAvg, TPSfMinAvg, TPSfUptAvg, TPSfWatAvg, TPSorbAvg, TPtoSOILAvg, TPtoVOLAvg, TranspAvg, UnsatMoistAvg, and UnsatZavg.
02066 { 02067 int ix, iy, cellLoc; 02068 02069 for(ix=1; ix<=s0; ix++) 02070 for(iy=1; iy<=s1; iy++) 02071 if(ON_MAP[cellLoc= T(ix,iy)]) { 02072 TotHeadAvg[cellLoc] = 0.0; 02073 HydRelDepPosNegAvg[cellLoc] = 0.0; 02074 SfWatAvg[cellLoc] = 0.0; 02075 UnsatZavg[cellLoc] = 0.0; 02076 UnsatMoistAvg[cellLoc] = 0.0; 02077 RainAvg[cellLoc] = 0.0; 02078 EvapAvg[cellLoc] = 0.0; 02079 TranspAvg[cellLoc] = 0.0; 02080 ETAvg[cellLoc] = 0.0; 02081 SF_WT_VEL_magAvg[cellLoc] = 0.0; 02082 BCmodel_sfwatAvg[cellLoc] = 0.0; 02083 02084 TPSfWatAvg[cellLoc] = 0.0; 02085 TPSfUptAvg[cellLoc] = 0.0; 02086 TPSfMinAvg[cellLoc] = 0.0; 02087 TP_settlAvg[cellLoc] = 0.0; 02088 TPSedWatAvg[cellLoc] = 0.0; 02089 TPSedUptAvg[cellLoc] = 0.0; 02090 TPSedMinAvg[cellLoc] = 0.0; 02091 TPSorbAvg[cellLoc] = 0.0; 02092 TPtoVOLAvg[cellLoc] = 0.0; 02093 TPtoSOILAvg[cellLoc] = 0.0; 02094 02095 SaltSfAvg[cellLoc] = 0.0; 02096 SaltSedAvg[cellLoc] = 0.0; 02097 02098 Floc_fr_phBioAvg[cellLoc] = 0.0; 02099 02100 NC_PeriAvg[cellLoc] = 0.0; 02101 NC_Peri_nppAvg[cellLoc] = 0.0; 02102 NC_Peri_mortAvg[cellLoc] = 0.0; 02103 NC_PeriRespAvg[cellLoc] = 0.0; 02104 NC_PeriNutCFAvg[cellLoc] = 0.0; 02105 C_Peri_PCAvg[cellLoc] = 0.0; 02106 C_PeriAvg[cellLoc] = 0.0; 02107 C_Peri_nppAvg[cellLoc] = 0.0; 02108 C_Peri_mortAvg[cellLoc] = 0.0; 02109 C_PeriRespAvg[cellLoc] = 0.0; 02110 C_PeriNutCFAvg[cellLoc] = 0.0; 02111 C_Peri_PCAvg[cellLoc] = 0.0; 02112 PeriAvg[cellLoc] = 0.0; 02113 PeriLiteCFAvg[cellLoc] = 0.0; 02114 02115 Mac_nphBioAvg[cellLoc] = 0.0; 02116 Mac_phBioAvg[cellLoc] = 0.0; 02117 Mac_totBioAvg[cellLoc] = 0.0; 02118 Mac_nppAvg[cellLoc] = 0.0; 02119 Mac_nphMortAvg[cellLoc] = 0.0; 02120 Mac_phMortAvg[cellLoc] = 0.0; 02121 LAI_effAvg[cellLoc] = 0.0; 02122 Manning_nAvg[cellLoc] = 0.0; 02123 MacNutCfAvg[cellLoc] = 0.0; 02124 MacWatCfAvg[cellLoc] = 0.0; 02125 mac_nph_PCAvg[cellLoc] = 0.0; 02126 mac_ph_PCAvg[cellLoc] = 0.0; 02127 02128 SedElevAvg[cellLoc] = 0.0; 02129 } 02130 } /* end of Cell_reset_avg() */
| void Cell_reset_hydper | ( | void | ) |
Zero the array holding hydroperiod data in cells (after printing).
Definition at line 2050 of file BudgStats.c.
References HydPerAnn, ON_MAP, s0, s1, and T.
02051 { 02052 int ix, iy, cellLoc; 02053 02054 for(ix=1; ix<=s0; ix++) 02055 for(iy=1; iy<=s1; iy++) 02056 if(ON_MAP[cellLoc= T(ix,iy)]) { 02057 HydPerAnn[cellLoc] = 0.0; 02058 } 02059 }
| void Flux_SWater | ( | int | it, | |
| float * | SURFACE_WAT, | |||
| float * | SED_ELEV, | |||
| float * | HYD_MANNINGS_N, | |||
| double * | STUF1, | |||
| double * | STUF2, | |||
| double * | STUF3, | |||
| float * | SfWat_vel_day | |||
| ) |
Surface water horizontal flux.
This is the alternating direction function that first fluxes water in the E/W direction and then in the N/S direction. It sweeps from left to right, then goes back from right to left, then goes from top to bottom and finally from bottom to top. This alternates every hyd_iter.
Surface water model boundary exchanges may only occur across cells designated with designated attribute in the boundary condition map file.
The surface water variable is actually updated in the Flux_SWstuff function
- Parameters:
-
it Horizontal iteration number SURFACE_WAT SURFACE_WAT SED_ELEV SED_ELEV HYD_MANNINGS_N HYD_MANNINGS_N STUF1 SALT_SURF_WT STUF2 DINdummy STUF3 TP_SF_WT SfWat_vel_day SF_WT_VEL_mag
- Remarks:
- Surface water flow - levee interaction rules:
ON_MAP value: if running managed canal network, ON_MAP is modified in WatMgmt.c, beyond just in/out (true/false) of model active domain, to these values (expressed in integer format):- 0 Not in model active domain
- 1 Allow flow in no direction
- 2 Allow flow to east<->west
- 3 Allow flow to south<->north
- 4 Allow flow in all directions
- 101 water control structure interaction (allow no flow)
- 102 water control structure interaction (allow flow to east<->west)
- 103 water control structure interaction (allow flow to south<->north)
- 104 water control structure interaction (allow all flow)
- 201 water control structure interaction (allow no flow)
- 202 water control structure interaction (allow flow to east<->west)
- 203 water control structure interaction (allow flow to south<->north)
- 204 water control structure interaction (allow all flow) etc.
- (ON_MAP) (ON_MAP-1) (ON_MAP-1)&1 (ON_MAP-1)&2
- \000 -1 1 2
- \001 0 0 0
- \002 1 1 0
- \003 2 0 2
- \004 3 1 2
Boundary condition flow allowances (read from BoundCond.BIN input map, values all within "ON_MAP" model domain):- Value of (integer) BCondFlow:
- 1 Allow no flows external to model domain
- 3 Allow surface- and ground- water flows to/from external boundary cells
- 4 Allow groundwater (but not surface) flows to/from external boundary cells
- (9) Archaic, will remove (allowing groundwater flows, with static external stage conditions)
- Variables local to function
- ix, iy Model grid cell row, column, respectively
FFlux Water flux (m) between grid cells
Definition at line 121 of file Fluxes.c.
References BCondFlow, Flux_SWcells(), Flux_SWstuff(), ON_MAP, s0, s1, and T.
00123 { 00129 int ix, iy; 00130 float FFlux; 00131 00132 00133 00134 /* check the donor and recipients cells for a) on-map, b) the cell attribute that allows sfwater 00135 boundary flow from the system and c) the attribute that indicates levee presence: 00136 the levee attribute of 1 (bitwise) allows flow to east; 00137 attribute of 2 (bitwise) allows flow to south (levee atts calc'd by WatMgmt.c) 00138 */ 00139 00140 /* as always, x is row, y is column! */ 00141 for(ix=1; ix<=s0; ix++) 00142 { 00143 if (it%2) /* alternate loop directions every other hyd_iter (it) */ 00144 { 00145 for(iy=1; iy<=s1; iy++) /* loop from west to east */ 00146 { 00147 if( ( ON_MAP[T(ix,iy)] && ON_MAP[T(ix,iy+1)] && (int)(ON_MAP[T(ix,iy)]-1) & 1 ) || 00148 BCondFlow[T(ix,iy+1)] == 3 || BCondFlow[T(ix,iy)] == 3 ) 00149 { 00150 FFlux = Flux_SWcells(ix,iy,ix,iy+1,SURFACE_WAT,SED_ELEV,HYD_MANNINGS_N); 00151 /* FFlux units = m */ 00152 if (FFlux != 0.0) 00153 Flux_SWstuff ( ix,iy,ix,iy+1,FFlux,SURFACE_WAT,STUF1,STUF2,STUF3,SfWat_vel_day); 00154 00155 } /* endof if */ 00156 } /* end of for */ 00157 } /* end of if */ 00158 00159 00160 else 00161 { 00162 for(iy=s1; iy>=1; iy--) /* loop from east to west */ 00163 if( ( ON_MAP[T(ix,iy)] && ON_MAP[T(ix,iy-1)] && (int)(ON_MAP[T(ix,iy-1)]-1) & 1 ) || 00164 BCondFlow[T(ix,iy-1)] == 3 || BCondFlow[T(ix,iy)] == 3 ) 00165 { 00166 FFlux = Flux_SWcells(ix,iy-1,ix,iy,SURFACE_WAT,SED_ELEV,HYD_MANNINGS_N); 00167 /* FFlux units = m */ 00168 if (FFlux != 0.0) 00169 Flux_SWstuff ( ix,iy-1,ix,iy,FFlux,SURFACE_WAT,STUF1,STUF2,STUF3,SfWat_vel_day); 00170 00171 } /* end of if */ 00172 } /* end of else */ 00173 } 00174 00175 for(iy=1; iy<=s1; iy++) 00176 { 00177 if (it%2) /* alternate loop directions every other hyd_iter (it) */ 00178 { 00179 for(ix=1; ix<=s0; ix++) /* loop from north to south */ 00180 if( ( ON_MAP[T(ix,iy)] && ON_MAP[T(ix+1,iy)] && (int)(ON_MAP[T(ix,iy)]-1) & 2 ) || 00181 BCondFlow[T(ix+1,iy)] == 3 || BCondFlow[T(ix,iy)] == 3 ) 00182 { 00183 FFlux = Flux_SWcells(ix,iy,ix+1,iy,SURFACE_WAT,SED_ELEV,HYD_MANNINGS_N); 00184 /* FFlux units = m */ 00185 if (FFlux != 0.0) 00186 Flux_SWstuff ( ix,iy,ix+1,iy,FFlux,SURFACE_WAT,STUF1,STUF2,STUF3,SfWat_vel_day); 00187 } 00188 } 00189 00190 00191 else 00192 { 00193 for(ix=s0; ix>=1; ix--) /* loop from south to north */ 00194 if( ( ON_MAP[T(ix,iy)] && ON_MAP[T(ix-1,iy)] && (int)(ON_MAP[T(ix-1,iy)]-1) & 2 ) || 00195 BCondFlow[T(ix-1,iy)] == 3 || BCondFlow[T(ix,iy)] == 3 ) 00196 { 00197 00198 FFlux = Flux_SWcells(ix-1,iy,ix,iy,SURFACE_WAT,SED_ELEV,HYD_MANNINGS_N); 00199 /* FFlux units = m */ 00200 if (FFlux != 0.0) 00201 Flux_SWstuff ( ix-1,iy,ix,iy,FFlux,SURFACE_WAT,STUF1,STUF2,STUF3,SfWat_vel_day); 00202 00203 } /* end of if */ 00204 } /* end of else */ 00205 } /* end of for */ 00206 00207 } /* end of function */
| void Flux_GWater | ( | int | it, | |
| float * | SatWat, | |||
| float * | Unsat, | |||
| float * | SfWat, | |||
| float * | rate, | |||
| float * | poros, | |||
| float * | sp_yield, | |||
| float * | elev, | |||
| double * | gwSTUF1, | |||
| double * | gwSTUF2, | |||
| double * | gwSTUF3, | |||
| double * | swSTUF1, | |||
| double * | swSTUF2, | |||
| double * | swSTUF3 | |||
| ) |
Groundwater fluxing routine.
This is the alternating direction function that first fluxes water in the E/W direction and then in the N/S direction. It sweeps from left to right, then goes back from right to left, then goes from top to bottom and finally from bottom to top. This alternates every iteration. Water is fluxed with mass conservation, allowing outflow from (no inflow to) the model system.
- Parameters:
-
it Horizontal iteration number SatWat SAT_WATER Unsat UNSAT_WATER SfWat SURFACE_WAT rate HYD_RCCONDUCT poros HP_HYD_POROSITY sp_yield HP_HYD_SPEC_YIELD elev SED_ELEV gwSTUF1 SALT_SED_WT gwSTUF2 DINdummy gwSTUF3 TP_SED_WT swSTUF1 SALT_SURF_WT swSTUF2 DINdummy swSTUF3 TP_SF_WT
- Remarks:
- Groundwater fluxing (this function) is called every other horizontal hyd_iter (it = int(hyd_iter-1)/2 ), or half as frequently as the surface water flow calculations.
- 1 Allow no flows external to model domain
- 3 Allow surface- and ground- water flows to/from external boundary cells
- 4 Allow groundwater (but not surface) flows to/from external boundary cells
- (9) Archaic, will remove (allowing groundwater flows, with static external stage conditions)
Definition at line 721 of file Fluxes.c.
References BCondFlow, Flux_GWcells(), ON_MAP, s0, s1, and T.
00726 { 00727 int ix, iy; /* ix is row, iy is col! */ 00728 /* we only check the donor cell for on-map, allowing losses from the system */ 00729 for(ix=1; ix<=s0; ix++) 00730 { 00731 if (it%2) { /* alternate loop directions every other iteration (it = int(hyd_iter-1)/2 ) */ 00732 for(iy=1; iy<=s1; iy++) /* loop from west to east */ 00733 /* allow boundary flow if donor cell is marked 3 or higher (v2.5 had specific markings of 4 or 9, no longer pertinent in v2.6+) */ 00734 if (( ON_MAP[T(ix,iy)] && ON_MAP[T(ix,iy+1)]) || 00735 ((BCondFlow[T(ix,iy+1)]) > 2) || ((BCondFlow[T(ix,iy)]) > 2) ) 00736 { 00737 Flux_GWcells(ix,iy,ix,iy+1,SatWat, Unsat, SfWat, rate, poros, sp_yield, elev, 00738 gwSTUF1, gwSTUF2, gwSTUF3, swSTUF1, swSTUF2, swSTUF3); 00739 } 00740 } 00741 00742 else { 00743 for(iy=s1; iy>=1; iy--) /* loop from east to west */ 00744 /* allow boundary flow if donor cell is marked 3 or higher (v2.5 had specific markings of 4 or 9, no longer pertinent in v2.6+) */ 00745 if (( ON_MAP[T(ix,iy)] && ON_MAP[T(ix,iy-1)]) || 00746 ((BCondFlow[T(ix,iy-1)]) > 2) || ((BCondFlow[T(ix,iy)]) > 2) ) 00747 { 00748 00749 Flux_GWcells(ix,iy,ix,iy-1,SatWat, Unsat, SfWat, rate, poros, sp_yield, elev, 00750 gwSTUF1, gwSTUF2, gwSTUF3, swSTUF1, swSTUF2, swSTUF3); 00751 } 00752 } 00753 } 00754 00755 for(iy=1; iy<=s1; iy++) 00756 { 00757 if (it%2) { /* alternate loop directions every other iteration (it = int(hyd_iter-1)/2 ) */ 00758 for(ix=1; ix<=s0; ix++) /* loop from north to south */ 00759 /* allow boundary flow if donor cell is marked 3 or higher (v2.5 had specific markings of 4 or 9, no longer pertinent in v2.6+) */ 00760 if (( ON_MAP[T(ix,iy)] && ON_MAP[T(ix+1,iy)]) || 00761 ((BCondFlow[T(ix+1,iy)]) > 2) || ((BCondFlow[T(ix,iy)]) > 2) ) 00762 { 00763 00764 Flux_GWcells(ix,iy,ix+1,iy,SatWat, Unsat, SfWat, rate, poros, sp_yield, elev, 00765 gwSTUF1, gwSTUF2, gwSTUF3, swSTUF1, swSTUF2, swSTUF3); 00766 } 00767 } 00768 else { 00769 for(ix=s0; ix>=1; ix--) /* loop from south to north */ 00770 /* allow boundary flow if donor cell is marked 3 or higher (v2.5 had specific markings of 4 or 9, no longer pertinent in v2.6+) */ 00771 if (( ON_MAP[T(ix,iy)] && ON_MAP[T(ix-1,iy)]) || 00772 ((BCondFlow[T(ix-1,iy)]) > 2) || ((BCondFlow[T(ix,iy)]) > 2) ) 00773 { 00774 00775 Flux_GWcells(ix,iy,ix-1,iy,SatWat, Unsat, SfWat, rate, poros, sp_yield, elev, 00776 gwSTUF1, gwSTUF2, gwSTUF3, swSTUF1, swSTUF2, swSTUF3); 00777 } 00778 } 00779 } 00780 00781 }
| float* get_hab_parm | ( | char * | s_parm_name, | |
| int | s_parm_relval, | |||
| char * | parmName | |||
| ) |
Read the input data file to get a habitat-specfic parameter array.
This gets an array of the targeted parameter, a value for each of (usually) multiple habitats.
- Parameters:
-
s_parm_name Name of the sensitivity parameter being varied for this run s_parm_relval Indicator of current value of relative range in sensitivity parm values: nominal (0), low (1), or high (2) parmName String that identifies the parameter to be read in dbase file
- Returns:
- fTmp Array of values of single parameter across multiple habitats
Definition at line 671 of file Serial.c.
References dFile, EOL, EOS, False, get_Nth_parm(), gHabChar, goto_index(), habNumTot, match_Sparm(), MAX_NHAB, modelFileName, ModelPath, msgStr, ProgExec, ProjName, prog_attr::S_ParmVal, Scip(), TAB, True, usrErr(), and WriteMsg().
00672 { 00673 int hIndex, end=0; 00674 FILE* parmFile; 00675 float* fTmp; 00676 char lline[2001], *line; 00677 int pLoc = -1; 00678 int foundParm= False; 00679 char parmNameHead[30]; 00680 char modelFileName[300]; 00681 char HabParmFile[300]; 00682 char *fileAppend; 00683 extern ProgAttr *ProgExec; 00684 00685 00686 if ( (fTmp = (float*) malloc( MAX_NHAB * sizeof(float) ) ) == NULL) { 00687 sprintf(msgStr, "Failed to allocate memory for HabParm array.\n ") ; 00688 usrErr(msgStr); 00689 exit( -2 ) ; 00690 }; 00691 fTmp[0] = 0.0; 00692 00693 fileAppend = match_Sparm( s_parm_name, s_parm_relval, parmName); 00694 sprintf(HabParmFile,"HabParms_%s",fileAppend); 00695 sprintf(modelFileName,"%s/%s/Data/%s",ModelPath,ProjName,HabParmFile); 00696 parmFile = fopen(modelFileName,"r"); 00697 if(parmFile==NULL) {fprintf(stdout,"\nERROR: Unable to open dBase file %s\n",modelFileName); exit(-1); } 00698 00699 fgets(lline, 2000, parmFile); /* skip 1st header line */ 00700 fgets(lline, 2000, parmFile); /* read 2nd header line with column names */ 00701 line=lline; 00702 00703 while (!foundParm) 00704 { 00705 sscanf( line,"%s",&parmNameHead); 00706 if (strcmp(parmName,parmNameHead) ==0) foundParm = True; 00707 pLoc++; 00708 if (*line == EOS || *line == EOL) { 00709 sprintf(msgStr,"ERROR: Could not find header tag of %s in HabParms dbase file.", parmName); 00710 usrErr(msgStr); 00711 exit(-1); 00712 } 00713 line = Scip( line, TAB ); 00714 } 00715 00716 sprintf(msgStr,"Parameter group: %s",parmName); 00717 fflush(dFile); 00718 WriteMsg(msgStr,1); 00719 for( hIndex = 1; hIndex < MAX_NHAB; hIndex++) { 00720 goto_index (parmFile, gHabChar, hIndex); 00721 fTmp[hIndex] = get_Nth_parm( parmFile, pLoc, &end, hIndex ); 00722 if(end) break; 00723 } 00724 fclose(parmFile); 00725 habNumTot = hIndex-1; 00726 00727 fflush(dFile); 00728 /* TODO: this HARDCODES into the BIR (BIRavg) model output file ONLY, 00729 the incorrect impression of running sensitivity on a single habitat ID 00730 (hab 2 = sawgrass plain, ELMv2.4) */ 00731 if (strcmp(s_parm_name,parmName) == 0) ProgExec->S_ParmVal = fTmp[2]; 00732 return fTmp; 00733 }
| float get_global_parm | ( | char * | s_parm_name, | |
| int | s_parm_relval, | |||
| char * | parmName | |||
| ) |
Read the input data file to get a global parameter.
This gets the single global value of the targeted parameter.
- Parameters:
-
s_parm_name Name of the sensitivity parameter being varied for this run s_parm_relval Indicator of current value of relative range in sensitivity parm values: nominal (0), low (1), or high (2) parmName String that identifies the parameter to be read in dbase file
- Returns:
- parmVal The value of the global parameter
Definition at line 775 of file Serial.c.
References Exit(), match_Sparm(), modelFileName, ModelPath, msgStr, ProgExec, ProjName, prog_attr::S_ParmVal, scan_forward(), usrErr(), and WriteMsg().
00776 { 00777 int test; 00778 char modelFileName[300]; 00779 FILE *parmFile; 00780 float parmVal; 00781 char parmNameHead[30]; 00782 extern ProgAttr *ProgExec; 00783 00784 00785 char GlobalParmFile[50]; 00786 char *fileAppend; 00787 00788 fileAppend = match_Sparm( s_parm_name, s_parm_relval, parmName); 00789 sprintf(GlobalParmFile,"GlobalParms_%s",fileAppend); 00790 sprintf(modelFileName,"%s/%s/Data/%s",ModelPath,ProjName,GlobalParmFile); 00791 00792 parmFile = fopen(modelFileName,"r"); 00793 if(parmFile==NULL) { 00794 sprintf(msgStr,"ERROR, can't open data file %s",modelFileName); 00795 usrErr(msgStr); 00796 Exit(0); 00797 } 00798 00799 sprintf(parmNameHead,"%s=", parmName); 00800 scan_forward(parmFile,parmNameHead); 00801 if ( (test = fscanf(parmFile,"%f",&parmVal) ) <= 0 ) { 00802 sprintf(msgStr,"ERROR in reading %s from %s; see Driver0.out debug file.", parmName, modelFileName); 00803 usrErr(msgStr); 00804 Exit(0); 00805 } 00806 sprintf(msgStr,"%s %f\n",parmNameHead, parmVal); 00807 WriteMsg(msgStr,1); 00808 00809 fclose(parmFile); 00810 if (strcmp(s_parm_name,parmName) == 0) ProgExec->S_ParmVal = parmVal; 00811 return parmVal; 00812 }
| float get_modexperim_parm | ( | char * | s_parm_name, | |
| int | s_parm_relval, | |||
| char * | parmName | |||
| ) |
Read the input data file to get a model experiment parameter.
This gets the single global value of the targeted parameter for special model experiments. (Added for ELM v2.6).
- Parameters:
-
s_parm_name Name of the sensitivity parameter being varied for this run s_parm_relval Indicator of current value of relative range in sensitivity parm values: nominal (0), low (1), or high (2) parmName String that identifies the parameter to be read in dbase file
- Returns:
- parmVal The value of the (global) model experiment parameter
Definition at line 822 of file Serial.c.
References Exit(), match_Sparm(), modelFileName, ModelPath, msgStr, ProgExec, ProjName, prog_attr::S_ParmVal, scan_forward(), usrErr(), and WriteMsg().
00823 { 00824 int test; 00825 char modelFileName[300]; 00826 FILE *parmFile; 00827 float parmVal; 00828 char parmNameHead[30]; 00829 extern ProgAttr *ProgExec; 00830 00831 00832 char ModExParmFile[50]; 00833 char *fileAppend; 00834 00835 fileAppend = match_Sparm( s_parm_name, s_parm_relval, parmName); 00836 sprintf(ModExParmFile,"ModExperimParms_%s",fileAppend); 00837 sprintf(modelFileName,"%s/%s/Data/%s",ModelPath,ProjName,ModExParmFile); 00838 00839 parmFile = fopen(modelFileName,"r"); 00840 if(parmFile==NULL) { 00841 sprintf(msgStr,"ERROR, can't open data file %s",modelFileName); 00842 usrErr(msgStr); 00843 Exit(0); 00844 } 00845 00846 sprintf(parmNameHead,"%s=", parmName); 00847 scan_forward(parmFile,parmNameHead); 00848 if ( (test = fscanf(parmFile,"%f",&parmVal) ) <= 0 ) { 00849 sprintf(msgStr,"ERROR in reading %s from %s; see Driver0.out debug file.", parmName, modelFileName); 00850 usrErr(msgStr); 00851 Exit(0); 00852 } 00853 sprintf(msgStr,"%s %f\n",parmNameHead, parmVal); 00854 WriteMsg(msgStr,1); 00855 00856 fclose(parmFile); 00857 if (strcmp(s_parm_name,parmName) == 0) ProgExec->S_ParmVal = parmVal; 00858 return parmVal; 00859 }
| int stage_data_wmm | ( | float * | ) |
stage (depth) data array
Definition at line 31 of file stage_inp.c.
References conv_ftTOm, applicationStruct::dataELM, applicationStruct::day, debug, elm_OG_map, elm_wmm_map, initDataStruct(), mapGrids(), ModelPath, msgStr, printGridMap(), processData(), ProjName, applicationStruct::recRead, returnData(), s0, s1, SimTime, stage_binfilename, stage_struct, T, simTime::TIME, and usrErr().
00032 { 00033 00034 int i,j,k; 00035 int success = 1, fail = -1; 00036 int stat = success; 00037 char gridmapfilename[335]; 00038 00039 if(SimTime.TIME==0) { 00040 /* elm_OG_map is data structure containing the mapping attributes at two scales */ 00041 elm_wmm_map = elm_OG_map; 00042 00043 if(elm_wmm_map == NULL) { 00044 sprintf(msgStr, "Mapping grids and setting up stage data..."); 00045 usrErr (msgStr); 00046 00047 sprintf(gridmapfilename, "%s%s/Data/gridmapping.txt", ModelPath, ProjName ); 00048 stat = mapGrids(gridmapfilename); 00049 elm_wmm_map = elm_OG_map; 00050 } 00051 00052 if(debug > 4) { 00053 printGridMap(); 00054 sprintf(msgStr,"stage_data_wmm==> Finished mapping grids"); 00055 usrErr (msgStr); 00056 } 00057 00058 sprintf(stage_binfilename, "%s%s/Data/BoundCond_stage.BIN", ModelPath, ProjName ); 00059 /* initializing data structures, move pointer to initial date (gridmap.c) */ 00060 stat = initDataStruct(stage_binfilename,&stage_struct); 00061 00062 if(debug > 4) { 00063 /*printELM2Grid_io(); */ 00064 /*drawELM2Grid_io(); */ 00065 sprintf(msgStr,"stage_data_wmm==> Finished initializing"); 00066 usrErr (msgStr); 00067 } 00068 00069 } /* end of SimTime.TIME=0 */ 00070 00071 00072 if(stage_struct.day >= stage_struct.recRead) { /* process the data in batch */ 00073 sprintf(msgStr,"Processing batch of stage data..."); 00074 usrErr (msgStr); 00075 stat = processData(stage_binfilename,&stage_struct); 00076 00077 if(debug > 4 ) { 00078 /*printBatchData(stageWMM,gridio_batch_len,widCnt);*/ /* TODO: remove this printBatchData function when sure is no longer needed */ 00079 sprintf(msgStr,"stage_data_wmm==> Finished processing data"); 00080 usrErr (msgStr); 00081 } 00082 } /* end of if */ 00083 00084 00085 if(stage_struct.day < stage_struct.recRead) { /* pass the data day by day */ 00086 returnData(stageSME,&stage_struct); 00087 00088 for(i = 0; i < s0; i++) { 00089 for(j = 0; j < s1; j++) { 00090 k = i*s1+j; 00091 stageSME[T((i+1),(j+1))] = stage_struct.dataELM[k] * conv_ftTOm; /* convert data from feet to meters */ 00092 } 00093 } 00094 00095 if(debug > 4) { 00096 sprintf(msgStr,"stage_data_wmm==> Finished returning data"); 00097 usrErr (msgStr); 00098 } 00099 00100 } /* end of if */ 00101 00102 return success; 00103 }
| int rain_data_wmm | ( | float * | ) |
rainfall data array
Definition at line 26 of file rain_inp.c.
References conv_inTOtenths_mm, applicationStruct::dataELM, applicationStruct::day, debug, elm_OG_map, elm_wmm_map, initDataStruct(), mapGrids(), ModelPath, msgStr, printGridMap(), processData(), ProjName, rain_binfilename, rain_struct, applicationStruct::recRead, returnData(), s0, s1, SimTime, T, simTime::TIME, usrErr(), and usrErr0().
00027 { 00028 00029 int i,j,k; 00030 int success = 1, fail = -1; 00031 int stat = success; 00032 char gridmapfilename[335]; 00033 00034 if(SimTime.TIME==0) { 00035 00036 // cleanUp(*elm_wmm_map, rainSME); 00037 00038 /* elm_OG_map is data structure containing the mapping attributes at two scales */ 00039 elm_wmm_map = elm_OG_map; 00040 00041 if(elm_wmm_map == NULL) { 00042 sprintf(msgStr, "Mapping grids and setting up rain data..."); 00043 usrErr0(msgStr); 00044 00045 sprintf(gridmapfilename, "%s%s/Data/gridmapping.txt", ModelPath, ProjName ); 00046 stat = mapGrids(gridmapfilename); 00047 elm_wmm_map = elm_OG_map; 00048 } 00049 00050 if(debug > 4) { 00051 printGridMap(); 00052 sprintf(msgStr,"rain_data_wmm==> Finished mapping grids"); 00053 usrErr (msgStr); 00054 } 00055 00056 sprintf(rain_binfilename, "%s%s/Data/rain.BIN", ModelPath, ProjName ); 00057 00058 /* initializing data structures, move pointer to initial date (gridmap.c) */ 00059 stat = initDataStruct(rain_binfilename,&rain_struct); 00060 00061 if(debug > 4) { 00062 /* printELM2Grid_io(); */ 00063 /*drawELM2Grid_io(); */ 00064 sprintf(msgStr,"rain_data_wmm==> Finished initializing"); 00065 usrErr (msgStr); 00066 } 00067 00068 } /* end of SimTime.TIME=0 */ 00069 00070 00071 if(rain_struct.day >= rain_struct.recRead) { /* process the data in batch */ 00072 sprintf(msgStr,"\nProcessing batch of rain data..."); 00073 usrErr (msgStr); 00074 stat = processData(rain_binfilename,&rain_struct); 00075 00076 if(debug > 4 ) { 00077 /*printBatchData(rainWMM,gridio_batch_len,widCnt);*/ /* TODO: remove this printBatchData function when sure is no longer needed */ 00078 sprintf(msgStr,"rain_data_wmm==> Finished processing data"); 00079 usrErr (msgStr); 00080 } 00081 } /* end of if */ 00082 00083 00084 if (rain_struct.day < rain_struct.recRead) { /* pass the data day by day */ 00085 returnData(rainSME,&rain_struct); 00086 /* change the unit here */ 00087 for(i = 0; i < s0; i++) { 00088 for(j = 0; j < s1; j++) { 00089 k = i*s1+j; 00090 rainSME[T((i+1),(j+1))] = rain_struct.dataELM[k] * conv_inTOtenths_mm; /* convert data from inches to tenths of mm */ 00091 } 00092 } 00093 00094 if(debug > 4) { 00095 sprintf(msgStr,"rain_data_wmm==> Finished returning data"); 00096 usrErr (msgStr); 00097 } 00098 00099 } /* end of if */ 00100 00101 return success; 00102 }
| int evap_data_wmm | ( | float * | ) |
potential ET data array
Definition at line 30 of file evap_inp.c.
References conv_inTOtenths_mm, applicationStruct::dataELM, applicationStruct::day, debug, elm_OG_map, elm_wmm_map, evap_binfilename, evap_struct, initDataStruct(), mapGrids(), ModelPath, msgStr, printGridMap(), processData(), ProjName, applicationStruct::recRead, returnData(), s0, s1, SimTime, T, simTime::TIME, and usrErr().
00031 { 00032 00033 int i,j,k; 00034 int success = 1, fail = -1; 00035 int stat = success; 00036 char gridmapfilename[335]; 00037 00038 if(SimTime.TIME==0) { 00039 /* elm_OG_map is data structure containing the mapping attributes at two scales */ 00040 elm_wmm_map = elm_OG_map; 00041 00042 if(elm_wmm_map == NULL) { 00043 sprintf(msgStr, "Mapping grids and setting up pET data..."); 00044 usrErr (msgStr); 00045 00046 sprintf(gridmapfilename, "%s%s/Data/gridmapping.txt", ModelPath, ProjName ); 00047 stat = mapGrids(gridmapfilename); 00048 elm_wmm_map = elm_OG_map; 00049 } 00050 00051 if(debug > 4) { 00052 printGridMap(); 00053 sprintf(msgStr,"evap_data_wmm==> Finished mapping grids"); 00054 usrErr (msgStr); 00055 } 00056 00057 sprintf(evap_binfilename, "%s%s/Data/ETp.BIN", ModelPath, ProjName ); 00058 /* initializing data structures, move pointer to initial date (gridmap.c) */ 00059 stat = initDataStruct(evap_binfilename,&evap_struct); 00060 00061 if(debug > 4) { 00062 /*printELM2Grid_io(); */ 00063 /*drawELM2Grid_io(); */ 00064 sprintf(msgStr,"evap_data_wmm==> Finished initializing"); 00065 usrErr (msgStr); 00066 } 00067 00068 } /* end of SimTime.TIME=0 */ 00069 00070 00071 if(evap_struct.day >= evap_struct.recRead) { /* process the data in batch */ 00072 sprintf(msgStr,"Processing batch of pET data..."); 00073 usrErr (msgStr); 00074 stat = processData(evap_binfilename,&evap_struct); 00075 00076 if(debug > 4 ) { 00077 /*printBatchData(evapWMM,gridio_batch_len,widCnt);*/ /* TODO: remove this printBatchData function when sure is no longer needed */ 00078 sprintf(msgStr,"evap_data_wmm==> Finished processing data"); 00079 usrErr (msgStr); 00080 } 00081 } /* end of if */ 00082 00083 00084 if(evap_struct.day < evap_struct.recRead) { /* pass the data day by day */ 00085 returnData(evapSME,&evap_struct); 00086 00087 for(i = 0; i < s0; i++) { 00088 for(j = 0; j < s1; j++) { 00089 k = i*s1+j; 00090 evapSME[T((i+1),(j+1))] = evap_struct.dataELM[k] * conv_inTOtenths_mm; /* convert data from inches to tenths of mm */ 00091 } 00092 } 00093 00094 if(debug > 4) { 00095 sprintf(msgStr,"evap_data_wmm==> Finished returning data"); 00096 usrErr (msgStr); 00097 } 00098 00099 } /* end of if */ 00100 00101 return success; 00102 }
Variable Documentation
| float DAYJUL |
A "julian" day counter within a 365 day "year"
Definition at line 31 of file unitmod.h.
Referenced by cell_dyn1(), CellAvg(), gen_output(), and init_eqns().
| float LATRAD |
- Non-spatial solar radiation intermediate variables.
- These (unmodified) complex calculations were developed, tested, and published for global applications by Nikolov & Zeller (1992).
LATRAD Regional latitude calculated in radians
SOLALPHA Intermediate calculation
SOLALTCORR Intermediate calculation (altitude correction)
SOLBETA Intermediate calculation
SOLCOSDEC Intermediate calculation
SOLDEC Intermediate calculation
SOLDEC1 Intermediate calculation
SOLELEV_SINE Intermediate calculation
SOLRADATMOS Solar radiation in upper atmosphere (cal/cm2/d)
SOLRISSET_HA Intermediate calculation
SOLRISSET_HA1 Intermediate calculation
Definition at line 49 of file unitmod.h.
Referenced by cell_dyn1(), and init_eqns().
| float SOLALPHA |
| float SOLALTCORR |
| float SOLBETA |
| float SOLCOSDEC |
| float SOLDEC |
| float SOLDEC1 |
| float SOLELEV_SINE |
| float SOLRADATMOS |
| float SOLRISSET_HA |
| float SOLRISSET_HA1 |
| float dispParm_scaled |
| float GP_BC_SfWat_add |
Model experiment (global) parameter. _Units_: m; _Brief_: ***add (pos or neg) value to Surface Water Depth in Boundary Condition cells outside model domain; _Extended_: used for very long time scale and/or very fine spatial scale subregional apps, where standard gridIO data are not useful
Definition at line 56 of file unitmod.h.
Referenced by Flux_SWcells(), and ReadModExperimParms().
| float GP_BC_SfWat_addHi |
Model experiment (global) parameter. _Units_: m; _Brief_: ***when SfWat > target, this depth value is used in non-linear fcn to add (pos or neg) value to Surface Water Depth in Boundary Condition cells outside model domain; _Extended_: used for very long time scale and/or very fine spatial scale subregional apps, where standard gridIO data are not useful
Definition at line 57 of file unitmod.h.
Referenced by Flux_SWcells(), and ReadModExperimParms().
| float GP_BC_SatWat_add |
Model experiment (global) parameter. _Units_: m; _Brief_: ***add (pos or neg) value to Saturated (ground) Water Depth in Boundary Condition cells outside model domain; _Extended_: used for very long time scale and/or very fine spatial scale subregional apps, where standard gridIO data are not useful
Definition at line 58 of file unitmod.h.
Referenced by Flux_GWcells(), and ReadModExperimParms().
| float GP_BC_SfWat_targ |
Model experiment (global) parameter. _Units_: m; _Brief_: ***maximum Surface Water Depth target in Boundary Condition cells inside model domain; _Extended_: used for very long time scale and/or very fine spatial scale subregional apps, where standard gridIO data are not useful
Definition at line 59 of file unitmod.h.
Referenced by Flux_SWcells(), and ReadModExperimParms().
| int GP_BC_InputRow |
Model experiment (global) parameter. _Units_: dimless; _Brief_: ***model grid row# below which (i.e., to north) the _SfWat_add parm is added, and above which (to south) the depth is subtracted; _Extended_: a "research" kluge for now - used for very long time scale and/or very fine spatial scale subregional apps, where standard gridIO data are not useful
Definition at line 60 of file unitmod.h.
Referenced by Flux_GWcells(), Flux_SWcells(), and ReadModExperimParms().
| float GP_BC_Pconc |
Model experiment (global) parameter. _Units_: ug/L; _Brief_: ***Phosphorus conc. in surface or ground water Boundary Condition inflows from outside model domain; _Extended_: new for v2.6
Definition at line 61 of file unitmod.h.
Referenced by Flux_SWstuff(), and ReadModExperimParms().
| float GP_BC_Sconc |
Model experiment (global) parameter. _Units_: g/L; _Brief_: ***Salt (tracer, usually chloride) conc. in surface or ground water Boundary Condition inflows from outside model domain; _Extended_: new for v2.6
Definition at line 62 of file unitmod.h.
Referenced by Flux_SWstuff(), and ReadModExperimParms().
| float g7[11][2] |
Initial value:
{ {0.0000000,0.0000000},{0.1000000,0.0200000},{0.2000000,0.0400000},{0.3000000,0.0700000},
{0.4000000,0.1250000},{0.5000000,0.2150000},{0.6000000,0.3450000},{0.7000000,0.5750000},
{0.8000000,0.8550000},{0.9000000,0.9850000},{1.0000000,1.0000000} }
Definition at line 66 of file unitmod.h.
Referenced by graph7().
| float g8[11][2] |
Initial value:
{ {0.0000000,0.0050000},{0.1000000,0.0100000},{0.2000000,0.0400000},{0.3000000,0.1000000},
{0.4000000,0.2800000},{0.5000000,0.7150000},{0.6000000,0.8650000},{0.7000000,0.9350000},
{0.8000000,0.9750000},{0.9000000,0.9950000},{1.0000000,1.0000000} }
Definition at line 70 of file unitmod.h.
Referenced by graph8().
| int CalMonOut |
The outstep interval (in Model.outList input file) used to denote output intervals of 1-calendar-month instead of selectable-julian-day
Definition at line 32 of file driver_utilities.h.
Referenced by gen_output(), and readViewParms().
| float Timestep2 = 1.0 |
| int NPtTS2 = 0 |
| int TIndex2 = 0 |
| float Timestep3 = 1.0 |
| int NPtTS3 = 0 |
| int TIndex3 = 0 |
| float Timestep4 = 1.0 |
| int NPtTS4 = 0 |
| int TIndex4 = 0 |
| float Timestep5 = 1.0 |
| int NPtTS5 = 0 |
| int TIndex5 = 0 |
| float Timestep6 = 1.0 |
| int NPtTS6 = 0 |
| int TIndex6 = 0 |
Basin/Indicator Region data
Definition at line 31 of file budgstats.h.
Basin/Indicator Region data
Definition at line 32 of file budgstats.h.
| int WatMgmtOn |
boolean flag indicating water management mode
Definition at line 34 of file generic_driver.h.
Referenced by Flux_SWstuff(), get_parmf(), horizFlow(), and main().
| int ESPmodeON |
boolean flag indicating Everglades Settling-of Phosphorus mode. A mode with all biol/chem (non-hydro) modules turned off, running only a net settling rate module that reproduces equations and data from the SFWMD's old Everglades Water Quality Model (EWQM). Done only for comparision to full ecological ELM - ESP doesn't work very well!!
Definition at line 31 of file generic_driver.h.
Referenced by BIRbudg_date(), BIRbudg_print(), BIRbudg_reset(), BIRbudg_sum(), BIRbudg_sumFinal(), BIRoutfiles(), get_parmf(), init_static_data(), and main().
| int HabSwitchOn |
boolean flag indicating habitat succession mode
Definition at line 35 of file generic_driver.h.
Referenced by cell_dyn1(), get_parmf(), and main().
| int avgPrint |
boolean flag to indicate if recurring-averages is to be printed at current time
Definition at line 81 of file generic_driver.h.
Referenced by CellAvg(), gen_output(), and track_time().
| char* OutputPath |
base pathname for all model output (user input)
Definition at line 42 of file driver_utilities.h.
| char * ProjName |
Definition at line 40 of file driver_utilities.h.
 1.5.6
1.5.6